Cellular reagents for diagnostics and synthetic biology
- PMID: 30110361
- PMCID: PMC6093680
- DOI: 10.1371/journal.pone.0201681
Cellular reagents for diagnostics and synthetic biology
Abstract
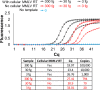
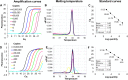
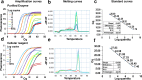
We have found that the overproduction of enzymes in bacteria followed by their lyophilization leads to 'cellular reagents' that can be directly used to carry out numerous molecular biology reactions. We demonstrate the use of cellular reagents in a variety of molecular diagnostics, such as TaqMan qPCR with no diminution in sensitivity, and in synthetic biology cornerstones such as the Gibson assembly of DNA fragments, where new plasmids can be constructed solely based on adding cellular reagents. Cellular reagents have significantly reduced complexity and cost of production, storage and implementation, features that should facilitate accessibility and use in resource-poor conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Treacy DJ, Sankaran SM, Gordon-Messer S, Saly D, Miller R, Isaac RS, et al. (2011) Implementation of a Project-Based Molecular Biology Laboratory Emphasizing Protein Structure-Function Relationships in a Large Introductory Biology Laboratory Course. Cbe-Life Sciences Education 10: 18–24. 10.1187/cbe.10-07-0085 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

