Large-Scale Human Dendritic Cell Differentiation Revealing Notch-Dependent Lineage Bifurcation and Heterogeneity
- PMID: 30110645
- PMCID: PMC6113934
- DOI: 10.1016/j.celrep.2018.07.033
Large-Scale Human Dendritic Cell Differentiation Revealing Notch-Dependent Lineage Bifurcation and Heterogeneity
Abstract
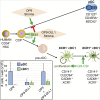
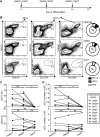
The ability to generate large numbers of distinct types of human dendritic cells (DCs) in vitro is critical for accelerating our understanding of DC biology and harnessing them clinically. We developed a DC differentiation method from human CD34+ precursors leading to high yields of plasmacytoid DCs (pDCs) and both types of conventional DCs (cDC1s and cDC2s). The identity of the cells generated in vitro and their strong homology to their blood counterparts were demonstrated by phenotypic, functional, and single-cell RNA-sequencing analyses. This culture system revealed a critical role of Notch signaling and GM-CSF for promoting cDC1 generation. Moreover, we discovered a pre-terminal differentiation state for each DC type, characterized by high expression of cell-cycle genes and lack of XCR1 in the case of cDC1. Our culture system will greatly facilitate the simultaneous and comprehensive study of primary, otherwise rare human DC types, including their mutual interactions.
Keywords: CLEC10A; CLEC9A; NOTCH; XCR1; adjuvant; dendritic cell differentiation; dendritic cell types; hematopoiesis; immunotherapy; plasmacytoid dendritic cells.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures






Comment in
-
'NOTCHing up' the In Vitro Production of Dendritic Cells.Trends Immunol. 2018 Oct;39(10):765-767. doi: 10.1016/j.it.2018.08.002. Epub 2018 Aug 24. Trends Immunol. 2018. PMID: 30150090
References
-
- Adema G.J., de Vries I.J., Punt C.J., Figdor C.G. Migration of dendritic cell based cancer vaccines: in vivo veritas? Curr. Opin. Immunol. 2005;17:170–174. - PubMed
-
- Bakdash G., Buschow S.I., Gorris M.A., Halilovic A., Hato S.V., Sköld A.E., Schreibelt G., Sittig S.P., Torensma R., Duiveman-de Boer T. Expansion of a BDCA1+CD14+ myeloid cell population in melanoma patients may attenuate the efficacy of dendritic cell vaccines. Cancer Res. 2016;76:4332–4346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

