New tools to screen wild peanut species for aflatoxin accumulation and genetic fingerprinting
- PMID: 30111278
- PMCID: PMC6094572
- DOI: 10.1186/s12870-018-1355-9
New tools to screen wild peanut species for aflatoxin accumulation and genetic fingerprinting
Abstract
Background: Aflatoxin contamination in peanut seeds is still a serious problem for the industry and human health. No stable aflatoxin resistant cultivars have yet been produced, and given the narrow genetic background of cultivated peanuts, wild species became an important source of genetic diversity. Wild peanut seeds, however, are not abundant, thus, an effective method of screening for aflatoxin accumulation using minimal seeds is highly desirable. In addition, keeping record of genetic fingerprinting of each accession would be very useful for breeding programs and for the identification of accessions within germplasm collections.
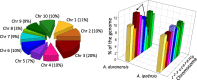
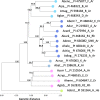
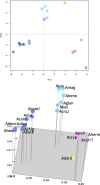
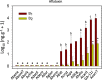
Results: In this study, we report a method of screening for aflatoxin accumulation that is applicable to the small-size seeds of wild peanuts, increases the reliability by testing seed viability, and records the genetic fingerprinting of the samples. Aflatoxin levels observed among 20 wild peanut species varied from zero to 19000 ng.g-1 and 155 ng.g-1 of aflatoxin B1 and B2, respectively. We report the screening of 373 molecular markers, including 288 novel SSRs, tested on 20 wild peanut species. Multivariate analysis by Neighbor-Joining, Principal Component Analysis and 3D-Principal Coordinate Analysis using 134 (36 %) transferable markers, in general grouped the samples according to their reported genomes. The best 88 markers, those with high fluorescence, good scorability and transferability, are reported with BLAST results. High quality markers (total 98) that discriminated genomes are reported. A high quality marker with UPIC score 16 (16 out of 20 species discriminated) had significant hits on BLAST2GO to a pentatricopeptide-repeat protein, another marker with score 5 had hits on UDP-D-apiose synthase, and a third one with score 12 had BLASTn hits on La-RP 1B protein. Together, these three markers discriminated all 20 species tested.
Conclusions: This study provides a reliable method to screen wild species of peanut for aflatoxin resistance using minimal seeds. In addition we report 288 new SSRs for peanut, and a cost-effective combination of markers sufficient to discriminate all 20 species tested. These tools can be used for the systematic search of aflatoxin resistant germplasm keeping record of the genetic fingerprinting of the accessions tested for breeding purpose.
Keywords: Arachis; Aspergillus flavus; Fingerprinting; aflatoxin; groundnut; molecular markers; peanut.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Blount WP. Turkey “X” disease. Turkeys. 1961;9:52–57.
-
- Mehan V, McDonald D, Ramakrishna N, Williams J. Effect of cultivar and date of harvest on infection of peanut seed by Aspergillus flavus and subsequent contamination with aflatoxin. Peanut Science. 1986;13:46–50. doi: 10.3146/i0095-3679-13-2-1. - DOI
-
- Rao MJV, Upadhyaya HD, Mehan VK, Nigam SN, Mcdonald D, Reddy NS. Registration of Peanut Germplasm Icgv-88145 and Icgv-89104 Resistant to Seed Infection by Aspergillus flavus. Crop Science. 1995;35(6):1717–7.
-
- Waliyar F, Kumar KVK, Diallo M, Traore A, Mangala UN, Upadhyaya HD, Sudini H. Resistance to pre-harvest aflatoxin contamination in ICRISAT’s groundnut mini core collection. European J Plant Pathology. 2016;145(4):901–913. doi: 10.1007/s10658-016-0879-9. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

