Development of a quadruple qRT-PCR assay for simultaneous identification of highly and low pathogenic H7N9 avian influenza viruses and characterization against oseltamivir resistance
- PMID: 30111290
- PMCID: PMC6094886
- DOI: 10.1186/s12879-018-3302-7
Development of a quadruple qRT-PCR assay for simultaneous identification of highly and low pathogenic H7N9 avian influenza viruses and characterization against oseltamivir resistance
Abstract
Background: During the fifth wave of human H7N9 infections, a novel highly pathogenic (HP) H7N9 variant emerged with an insertion of multiple basic amino acids in the HA cleavage site. Moreover, a neuraminidase inhibitor (NAI) resistance (R292K in NA) mutation was found in H7N9 isolates from humans, poultry and the environment. In this study, we set out to develop and validate a multiplex quantitative reverse transcript polymerase chain reaction (qRT-PCR) to simultaneously detect the presence of H7N9 and further identify the HP and NAI-resistance mutations.
Methods: A quadruple qRT-PCR to simultaneously detect the presence of H7N9 and further identify the HP and NAI-resistance mutations was designed based on the analyses of the HA and NA genes of H7N9. This assay was further tested for specificity and sensitivity, and validated using clinical samples.
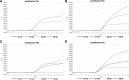
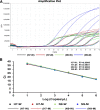
Results: The assay was highly specific and able to detect low pathogenic (LP)- or HP-H7N9 with/without the NAI-resistance mutation. The detection limit of the assay was determined to be 50 genome-equivalent copies and 2.8 × 10- 3 50% tissue culture infectious doses (TCID50) of live H7N9 per reaction. Clinical validation was confirmed by commercial kits and Sanger sequencing with ten clinical samples.
Conclusions: We developed and validated a rapid, single-reaction, one-step, quadruple real-time qRT-PCR to simultaneously detect the presence of H7N9 and further identify the HP- and NAI-resistance strains with excellent performance in specificity and sensitivity. This assay could be used to monitor the evolution of H7N9 viruses in the laboratory, field and the clinic for early-warning and the prevention of H7N9 infections.
Keywords: H7N9; Highly/low pathogenic avian influenza virus; Molecular diagnostics; NAI-resistance; Oseltamivir; Quadruple qRT-PCR.
Conflict of interest statement
Ethics approval and consent to participate
The study was performed in accordance with guidelines approved by the Ethics Committees from Shenzhen Third People’s Hospital (SZTHEC2016001) and Yunnan Center for Disease Control and Prevention Ethics Committee (YNCDC2017001), and verbal informed consents were obtained from all patients or patients’ family members.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
A novel pyrosequencing assay for the detection of neuraminidase inhibitor resistance-conferring mutations among clinical isolates of avian H7N9 influenza virus.Virus Res. 2014 Jan 22;179:119-24. doi: 10.1016/j.virusres.2013.10.026. Epub 2013 Nov 5. Virus Res. 2014. PMID: 24211668 Free PMC article.
-
Development of novel AllGlo-probe-based one-step multiplex qRT-PCR assay for rapid identification of avian influenza virus H7N9.Arch Virol. 2014 Jul;159(7):1707-13. doi: 10.1007/s00705-014-1979-5. Epub 2014 Jan 29. Arch Virol. 2014. PMID: 24473706
-
Neuraminidase Mutations Conferring Resistance to Oseltamivir in Influenza A(H7N9) Viruses.J Virol. 2015 May;89(10):5419-26. doi: 10.1128/JVI.03513-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740997 Free PMC article.
-
Research progress in human infection with avian influenza H7N9 virus.Sci China Life Sci. 2017 Dec;60(12):1299-1306. doi: 10.1007/s11427-017-9221-4. Epub 2017 Dec 1. Sci China Life Sci. 2017. PMID: 29270791 Review.
-
EMERGING PATHOGENS: INFLUENZA - H7N9.Dis Mon. 2017 Sep;63(9):251-256. doi: 10.1016/j.disamonth.2017.03.018. Epub 2017 Sep 12. Dis Mon. 2017. PMID: 29737282 Free PMC article. Review. No abstract available.
Cited by
-
Severe myocarditis due to influenza A(H1N1)pdm09 viral infection in a young woman successfully treated with intravenous zanamivir: A case report.Clin Case Rep. 2019 Oct 21;7(12):2336-2340. doi: 10.1002/ccr3.2499. eCollection 2019 Dec. Clin Case Rep. 2019. PMID: 31893053 Free PMC article.
-
Clustered Regularly Interspaced Short Palindromic Repeats-Associated Proteins13a combined with magnetic beads, chemiluminescence and reverse transcription-recombinase aided amplification for detection of avian influenza a (H7N9) virus.Front Bioeng Biotechnol. 2023 Jan 5;10:1094028. doi: 10.3389/fbioe.2022.1094028. eCollection 2022. Front Bioeng Biotechnol. 2023. PMID: 36686235 Free PMC article.
-
Dominant subtype switch in avian influenza viruses during 2016-2019 in China.Nat Commun. 2020 Nov 20;11(1):5909. doi: 10.1038/s41467-020-19671-3. Nat Commun. 2020. PMID: 33219213 Free PMC article.
-
Identification of candidate reference genes for qRT-PCR normalization studies of salinity stress and injury in Onchidium reevesii.PeerJ. 2019 Apr 26;7:e6834. doi: 10.7717/peerj.6834. eCollection 2019. PeerJ. 2019. PMID: 31086748 Free PMC article.
-
Longitudinal analysis of antibody dynamics in COVID-19 convalescents reveals neutralizing responses up to 16 months after infection.Nat Microbiol. 2022 Mar;7(3):423-433. doi: 10.1038/s41564-021-01051-2. Epub 2022 Feb 7. Nat Microbiol. 2022. PMID: 35132197
References
-
- Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, Wang L, Huo X, Xu K, Chen E, et al. Epidemiology of avian influenza a H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17(8):822–832. doi: 10.1016/S1473-3099(17)30323-7. - DOI - PMC - PubMed
-
- WHO. Monthly risk assessment summary. 2017. http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en/. Accessed 4 Nov 2017.
MeSH terms
Substances
Grants and funding
- 2016YFE0205800/Intramural special grant for influenza virus research from the Chinese Academy of Sciences/International
- 2016YFE0205800/National Key Research and Development Project of China/International
- 2016ZX10004222/National Science and Technology Major Project/International
- ZDSYS201504301534057/Sanming Project of Medicine in Shenzhen/International
- JCYJ20160427153238750/Shenzhen Science and Technology Research and Development Project/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

