Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease
- PMID: 30111768
- PMCID: PMC6093935
- DOI: 10.1038/s41467-018-05512-x
Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease
Erratum in
-
Author Correction: Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease.Nat Commun. 2018 Sep 18;9(1):3853. doi: 10.1038/s41467-018-06231-z. Nat Commun. 2018. PMID: 30228274 Free PMC article.
Abstract
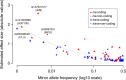
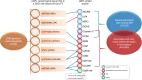
Identifying genetic variants associated with circulating protein concentrations (protein quantitative trait loci; pQTLs) and integrating them with variants from genome-wide association studies (GWAS) may illuminate the proteome's causal role in disease and bridge a knowledge gap regarding SNP-disease associations. We provide the results of GWAS of 71 high-value cardiovascular disease proteins in 6861 Framingham Heart Study participants and independent external replication. We report the mapping of over 16,000 pQTL variants and their functional relevance. We provide an integrated plasma protein-QTL database. Thirteen proteins harbor pQTL variants that match coronary disease-risk variants from GWAS or test causal for coronary disease by Mendelian randomization. Eight of these proteins predict new-onset cardiovascular disease events in Framingham participants. We demonstrate that identifying pQTLs, integrating them with GWAS results, employing Mendelian randomization, and prospectively testing protein-trait associations holds potential for elucidating causal genes, proteins, and pathways for cardiovascular disease and may identify targets for its prevention and treatment.
Conflict of interest statement
A.S.B. reports grants from Merck, Pfizer, Novartis, Biogen and Bioverativ and consulting fees from Novartis. J.D. sits on the Novartis Cardiovascular and Metabolic Advisory Board, and had grant support from Novartis. J.C.M. and H.R. were Merck employees at the time of their contributions to this study. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

