Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing
- PMID: 30111840
- PMCID: PMC6217926
- DOI: 10.1038/s41586-018-0423-5
Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing
Abstract
Common genetic contributions to autism spectrum disorder (ASD) reside in risk gene variants that individually have minimal effect sizes. As environmental factors that perturb neurodevelopment also underlie idiopathic ASD, it is crucial to identify altered regulators that can orchestrate multiple ASD risk genes during neurodevelopment. Cytoplasmic polyadenylation element binding proteins 1-4 (CPEB1-4) regulate the translation of specific mRNAs by modulating their poly(A)-tails and thereby participate in embryonic development and synaptic plasticity. Here we find that CPEB4 binds transcripts of most high-confidence ASD risk genes. The brains of individuals with idiopathic ASD show imbalances in CPEB4 transcript isoforms that result from decreased inclusion of a neuron-specific microexon. In addition, 9% of the transcriptome shows reduced poly(A)-tail length. Notably, this percentage is much higher for high-confidence ASD risk genes, correlating with reduced expression of the protein products of ASD risk genes. An equivalent imbalance in CPEB4 transcript isoforms in mice mimics the changes in mRNA polyadenylation and protein expression of ASD risk genes and induces ASD-like neuroanatomical, electrophysiological and behavioural phenotypes. Together, these data identify CPEB4 as a regulator of ASD risk genes.
Conflict of interest statement
Figures
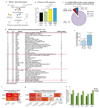

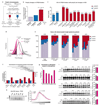
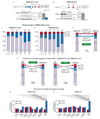




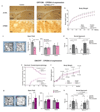



References
-
- Sztainberg Y, Zoghbi HY. Lessons learned from studying syndromic autism spectrum disorders. Nature neuroscience. 2016;19:1408–1417. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

