Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy
- PMID: 30111846
- PMCID: PMC6093931
- DOI: 10.1038/s41467-018-05763-8
Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy
Abstract
Triple-negative breast cancer (TNBC) lacks targeted therapies and has a worse prognosis than other breast cancer subtypes, underscoring an urgent need for new therapeutic targets and strategies. IRE1 is an endoplasmic reticulum (ER) stress sensor, whose activation is predominantly linked to the resolution of ER stress and, in the case of severe stress, to cell death. Here we demonstrate that constitutive IRE1 RNase activity contributes to basal production of pro-tumorigenic factors IL-6, IL-8, CXCL1, GM-CSF, and TGFβ2 in TNBC cells. We further show that the chemotherapeutic drug, paclitaxel, enhances IRE1 RNase activity and this contributes to paclitaxel-mediated expansion of tumor-initiating cells. In a xenograft mouse model of TNBC, inhibition of IRE1 RNase activity increases paclitaxel-mediated tumor suppression and delays tumor relapse post therapy. We therefore conclude that inclusion of IRE1 RNase inhibition in therapeutic strategies can enhance the effectiveness of current chemotherapeutics.
Conflict of interest statement
A.S., A.M.G., and E.C. are co-founders and shareholders of Cell Stress Discoveries Ltd. S.G., Q.Z., and J.B.P. are employees and shareholders of Fosun Orinove PharmaTech Inc. The remaining authors declare no competing interests.
Figures



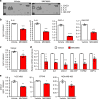

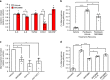


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

