PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing
- PMID: 30111894
- PMCID: PMC6105523
- DOI: 10.1038/s41590-018-0179-y
PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing
Abstract
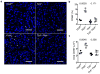
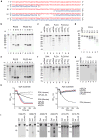
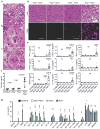
The sensing of microbial genetic material by leukocytes often elicits beneficial pro-inflammatory cytokines, but dysregulated responses can cause severe pathogenesis. Genome-wide association studies have linked the gene encoding phospholipase D3 (PLD3) to Alzheimer's disease and have linked PLD4 to rheumatoid arthritis and systemic sclerosis. PLD3 and PLD4 are endolysosomal proteins whose functions are obscure. Here, PLD4-deficient mice were found to have an inflammatory disease, marked by elevated levels of interferon-γ (IFN-γ) and splenomegaly. These phenotypes were traced to altered responsiveness of PLD4-deficient dendritic cells to ligands of the single-stranded DNA sensor TLR9. Macrophages from PLD3-deficient mice also had exaggerated TLR9 responses. Although PLD4 and PLD3 were presumed to be phospholipases, we found that they are 5' exonucleases, probably identical to spleen phosphodiesterase, that break down TLR9 ligands. Mice deficient in both PLD3 and PLD4 developed lethal liver inflammation in early life, which indicates that both enzymes are needed to regulate inflammatory cytokine responses via the degradation of nucleic acids.
Conflict of interest statement
The authors have no competing interests
Figures




References
-
- Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29:185–214. - PubMed
-
- Rigby RE, Leitch A, Jackson AP. Nucleic acid-mediated inflammatory diseases. Bioessays. 2008;30:833–842. - PubMed
-
- Al-Mayouf SM, et al. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat Genet. 2011;43:1186–1188. - PubMed
-
- Napirei M, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

