Comprehensive integrated analysis of gene expression datasets identifies key anti-cancer targets in different stages of breast cancer
- PMID: 30112036
- PMCID: PMC6090421
- DOI: 10.3892/etm.2018.6268
Comprehensive integrated analysis of gene expression datasets identifies key anti-cancer targets in different stages of breast cancer
Abstract
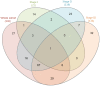
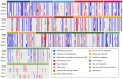
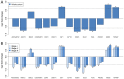
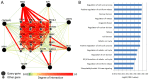
Breast cancer is one of the primary threats to women's health worldwide. However, the molecular mechanisms underlying the development of breast cancer remain to be fully elucidated. The present study aimed to investigate specific target gene expression profiles in breast cancer tissues in general and in different breast cancer stages, as well as to explore their functions in tumor development. For integrated analysis, a total of 5 gene expression profiling datasets for 3 different stages of breast cancer (stages I-III) were downloaded from the Gene Expression Omnibus of the National Center for Biotechnology Information. Pre-processing of these datasets was performed using the Robust Multi-array Average algorithm and global renormalization was performed for all studies. Differentially expressed genes between breast cancer patients and controls were estimated using the empirical Bayes algorithm. The Database for Annotation, Visualization and Integrated Discovery web server was used for analyzing the enrichment of the differentially expressed genes in Gene Ontology terms of the category biological process and in Kyoto Encyclopedia of Genes and Genomes pathways. Furthermore, breast cancer target genes were downloaded from the Thomson Reuters Integrity Database. We merged these target genes with the genes in breast cancer datasets. Analysis of anti-breast cancer gene networks was performed using the Genome-scale Integrated Analysis of Gene Networks in Tissues web server. The results demonstrated that the normal functions of the cell cycle, cell migration and cell adhesion were altered in all stages of breast cancer. Furthermore, 12 anti-breast cancer genes were identified to be dysregulated in at least one of the three stages. Among all of these genes, ribonucleotide reductase regulatory subunit M2 (RRM2) exhibited the highest degree of interaction with other interacting genes. Analysis of the network interactions revealed that the transcription factor of RRM2 is crucial for cancer development. Other genes, including mucin 1, progesterone receptor and cyclin-dependent kinase 5 regulatory subunit associated protein 3, also exhibited a high degree of interaction with the associated genes. In conclusion, several key anti-breast cancer genes identified in the present study are mainly associated with the regulation of the cell cycle, cell migration, cell adhesion and other cancer-associated cell functions, particularly RRM2.
Keywords: breast cancer; gene expression; network; ribonucleotide reductase regulatory subunit M2; target genes.
Figures
Similar articles
-
Identifying the key genes and microRNAs in colorectal cancer liver metastasis by bioinformatics analysis and in vitro experiments.Oncol Rep. 2019 Jan;41(1):279-291. doi: 10.3892/or.2018.6840. Epub 2018 Nov 1. Oncol Rep. 2019. PMID: 30542696 Free PMC article.
-
High-efficient Screening Method for Identification of Key Genes in Breast Cancer Through Microarray and Bioinformatics.Anticancer Res. 2017 Aug;37(8):4329-4335. doi: 10.21873/anticanres.11826. Anticancer Res. 2017. PMID: 28739725
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
-
Evaluation of core serous epithelial ovarian cancer genes as potential prognostic markers and indicators of the underlying molecular mechanisms using an integrated bioinformatics analysis.Oncol Lett. 2019 Nov;18(5):5508-5522. doi: 10.3892/ol.2019.10884. Epub 2019 Sep 19. Oncol Lett. 2019. PMID: 31612059 Free PMC article.
-
Functional Signatures in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis of Sex-Based Differences in Transcriptomic Studies.Cancers (Basel). 2021 Jan 5;13(1):143. doi: 10.3390/cancers13010143. Cancers (Basel). 2021. PMID: 33526761 Free PMC article. Review.
Cited by
-
SCIPAC: quantitative estimation of cell-phenotype associations.Genome Biol. 2024 May 13;25(1):119. doi: 10.1186/s13059-024-03263-1. Genome Biol. 2024. PMID: 38741183 Free PMC article.
-
Key Genes and Prognostic Analysis in HER2+ Breast Cancer.Technol Cancer Res Treat. 2021 Jan-Dec;20:1533033820983298. doi: 10.1177/1533033820983298. Technol Cancer Res Treat. 2021. PMID: 33499770 Free PMC article.
-
A systematic review of datasets that can help elucidate relationships among gene expression, race, and immunohistochemistry-defined subtypes in breast cancer.Cancer Biol Ther. 2021 Sep 2;22(7-9):417-429. doi: 10.1080/15384047.2021.1953902. Epub 2021 Aug 19. Cancer Biol Ther. 2021. PMID: 34412551 Free PMC article.
-
Enhanced detection of circulating tumor cells using a MUC1 promoter-driven recombinant adenovirus.Front Oncol. 2025 Jan 16;14:1506968. doi: 10.3389/fonc.2024.1506968. eCollection 2024. Front Oncol. 2025. PMID: 39886667 Free PMC article.
References
-
- Global Burden of Disease Cancer Collaboration, corp-author. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
