Discovery and genome characterization of three new Jeilongviruses, a lineage of paramyxoviruses characterized by their unique membrane proteins
- PMID: 30115009
- PMCID: PMC6097224
- DOI: 10.1186/s12864-018-4995-0
Discovery and genome characterization of three new Jeilongviruses, a lineage of paramyxoviruses characterized by their unique membrane proteins
Abstract
Background: In the past decade, many new paramyxoviruses that do not belong to any of the seven established genera in the family Paramyxoviridae have been discovered. Amongst them are J-virus (JPV), Beilong virus (BeiPV) and Tailam virus (TlmPV), three paramyxovirus species found in rodents. Based on their similarities, it has been suggested that these viruses should compose a new genus, tentatively called 'Jeilongvirus'.
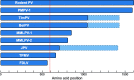
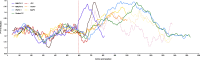
Results: Here we present the complete genomes of three newly discovered paramyxoviruses, one found in a bank vole (Myodes glareolus) from Slovenia and two in a single, co-infected Rungwe brush-furred rat (Lophuromys machangui) from Mozambique, that represent three new, separate species within the putative genus 'Jeilongvirus'. The genome organization of these viruses is similar to other paramyxoviruses, but like JPV, BeiPV and TlmPV, they possess an additional open reading frame, encoding a transmembrane protein, that is located between the F and G genes. As is the case for all Jeilongviruses, the G genes of the viruses described here are unusually large, and their encoded proteins are characterized by a remarkable amino acid composition pattern that is not seen in other paramyxoviruses, but resembles certain motifs found in Orthopneumovirus G proteins.
Conclusions: The phylogenetic clustering of JPV, BeiPV and TlmPV with the viruses described here, as well as their shared features that set them apart from other paramyxoviruses, provide additional support for the recognition of the genus 'Jeilongvirus'.
Keywords: Cell attachment protein; G protein; MMLPV-1; MMLPV-2; PMPV-1; Rodent paramyxovirus.
Conflict of interest statement
Ethics approval and consent to participate
Small mammal sampling in Slovenia was done in cooperation with the Slovenian Museum of Natural History. For the trappings in Mozambique, a small mammal-sampling permit was acquired from the National Directorate for Protected Areas (DINAC – Mozambique). All animal work was approved either by the Administration of the Republic of Slovenia for Food Safety, Veterinary Sector and Plant Protection (U34401–20/2013/5) or by the University of Antwerp Ethical Committee for Animal Experimentation (2011–52).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Samal SK. The biology of paramyxoviruses. Norfolk, UK: Caister Academic Press; 2011.
MeSH terms
Substances
Grants and funding
- 1S28617N/Fonds Wetenschappelijk Onderzoek
- 1S61618N/Fonds Wetenschappelijk Onderzoek
- 12T1117N/Fonds Wetenschappelijk Onderzoek
- 1167112N/Fonds Wetenschappelijk Onderzoek
- 12U7118N/Fonds Wetenschappelijk Onderzoek
- G066215N/Fonds Wetenschappelijk Onderzoek
- G0D5117N/Fonds Wetenschappelijk Onderzoek
- G0B9317N/Fonds Wetenschappelijk Onderzoek
- P3-0083/Javna Agencija za Raziskovalno Dejavnost RS
- P506-10-0983/Grantová Agentura České Republiky
- 653316/H2020 European Research Council
- 725422/H2020 European Research Council
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

