Optical and physical mapping with local finishing enables megabase-scale resolution of agronomically important regions in the wheat genome
- PMID: 30115128
- PMCID: PMC6097218
- DOI: 10.1186/s13059-018-1475-4
Optical and physical mapping with local finishing enables megabase-scale resolution of agronomically important regions in the wheat genome
Abstract
Background: Numerous scaffold-level sequences for wheat are now being released and, in this context, we report on a strategy for improving the overall assembly to a level comparable to that of the human genome.
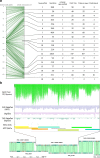
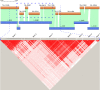
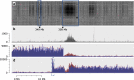
Results: Using chromosome 7A of wheat as a model, sequence-finished megabase-scale sections of this chromosome were established by combining a new independent assembly using a bacterial artificial chromosome (BAC)-based physical map, BAC pool paired-end sequencing, chromosome-arm-specific mate-pair sequencing and Bionano optical mapping with the International Wheat Genome Sequencing Consortium RefSeq v1.0 sequence and its underlying raw data. The combined assembly results in 18 super-scaffolds across the chromosome. The value of finished genome regions is demonstrated for two approximately 2.5 Mb regions associated with yield and the grain quality phenotype of fructan carbohydrate grain levels. In addition, the 50 Mb centromere region analysis incorporates cytological data highlighting the importance of non-sequence data in the assembly of this complex genome region.
Conclusions: Sufficient genome sequence information is shown to now be available for the wheat community to produce sequence-finished releases of each chromosome of the reference genome. The high-level completion identified that an array of seven fructosyl transferase genes underpins grain quality and that yield attributes are affected by five F-box-only-protein-ubiquitin ligase domain and four root-specific lipid transfer domain genes. The completed sequence also includes the centromere.
Keywords: Megabase-scale integration; Optical/physical maps Grain quality; Wheat sequence finishing; Yield.
Conflict of interest statement
Competing interests
PR, SB, and M-AN have competing commercial interests as employees and stockholders of Gydle, which is a commercial company that provides bioinformatics analysis software and services. This does not alter the authors’ adherence to all of the Genome Biology policies on sharing data and materials. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- The International Wheat Genome Sequencing Conosrtium. Shifting the limits in wheat research and breeding through a fully annotated and anchored reference genome sequence. Science. 2018. 10.1126/science.aar7191. - PubMed
-
- Clavijo BP, Kettleborough G, Heavens D, Chapman H, Lipscombe J, Barker T, Lu F-H, McKenzie N, Raats D, Ramirez-Gonzalez RH, Coince A, Peel N, Percival-Alwyn L, Duncan O, Trösch J, Yu G, Bolser DM, Namaati G, Kerhornou A, Spannagl M, Gundlach H, Haberer G, Davey RP, Fosker C, Di Palma FD, Phillips AL, Millar AH, Kersey PJ, Uauy C, Krasileva KW, Swarbreck D, Bevan MW, Clark MD. An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017;27:885–896. doi: 10.1101/gr.217117.116. - DOI - PMC - PubMed
-
- Eversole K, Rogers J, Keller B, Appels R, Feuillet C. Achieving sustainable cultivation of wheat, Part 1, Chap. 2. Cambridge: Burleigh-Dodds Science Publishing; 2017. Sequencing and assembly of the wheat genome.
Publication types
MeSH terms
Substances
Grants and funding
- UMU00037/Grains Research and Development Corporation/International
- ACSRF00542/Department of Industry, Innovation, Science, Research and Tertiary Education, Australian Government/International
- NA/Commonwealth Scientific and Industrial Research Organisation/International
- NA/BioPlatform Australia (BPA)/International
- NA/Victorian Department of Economic Development, Jobs, Transport and Resources/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

