Computational modeling and confirmation of leukemia-associated minor histocompatibility antigens
- PMID: 30115642
- PMCID: PMC6113610
- DOI: 10.1182/bloodadvances.2018022475
Computational modeling and confirmation of leukemia-associated minor histocompatibility antigens
Abstract
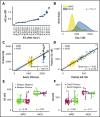
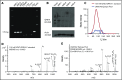
T-cell responses to minor histocompatibility antigens (mHAs) mediate both antitumor immunity (graft-versus-leukemia [GVL]) and graft-versus-host disease (GVHD) in allogeneic stem cell transplant. Identifying mHAs with high allele frequency, tight binding affinity to common HLA molecules, and narrow tissue restriction could enhance immunotherapy against leukemia. Genotyping and HLA allele data from 101 HLA-matched donor-recipient pairs (DRPs) were computationally analyzed to predict both class I and class II mHAs likely to induce either GVL or GVHD. Roughly twice as many mHAs were predicted in HLA-matched unrelated donor (MUD) stem cell transplantation (SCT) compared with HLA-matched related transplants, an expected result given greater genetic disparity in MUD SCT. Computational analysis predicted 14 of 18 previously identified mHAs, with 2 minor antigen mismatches not being contained in the patient cohort, 1 missed mHA resulting from a noncanonical translation of the peptide antigen, and 1 case of poor binding prediction. A predicted peptide epitope derived from GRK4, a protein expressed in acute myeloid leukemia and testis, was confirmed by targeted differential ion mobility spectrometry-tandem mass spectrometry. T cells specific to UNC-GRK4-V were identified by tetramer analysis both in DRPs where a minor antigen mismatch was predicted and in DRPs where the donor contained the allele encoding UNC-GRK4-V, suggesting that this antigen could be both an mHA and a cancer-testis antigen. Computational analysis of genomic and transcriptomic data can reliably predict leukemia-associated mHA and can be used to guide targeted mHA discovery.
© 2018 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: Bruker Daltonics has licensed, from the University of North Carolina, some intellectual property related to the DIMS technology that was developed in the Glish (G.L.G.) laboratory. The remaining authors declare no competing financial interests.
Figures





References
-
- Mullally A, Ritz J. Beyond HLA: the significance of genomic variation for allogeneic hematopoietic stem cell transplantation. Blood. 2007;109(4):1355-1362. - PubMed
-
- Gale RP, Horowitz MM, Ash RC, et al. . Identical-twin bone marrow transplants for leukemia. Ann Intern Med. 1994;120(8):646-652. - PubMed
-
- Horowitz MM, Gale RP, Sondel PM, et al. . Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75(3):555-562. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

