A Spatiotemporal DNA Endoploidy Map of the Arabidopsis Root Reveals Roles for the Endocycle in Root Development and Stress Adaptation
- PMID: 30115738
- PMCID: PMC6241279
- DOI: 10.1105/tpc.17.00983
A Spatiotemporal DNA Endoploidy Map of the Arabidopsis Root Reveals Roles for the Endocycle in Root Development and Stress Adaptation
Abstract
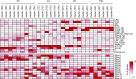
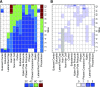
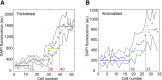
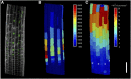
Somatic polyploidy caused by endoreplication is observed in arthropods, molluscs, and vertebrates but is especially prominent in higher plants, where it has been postulated to be essential for cell growth and fate maintenance. However, a comprehensive understanding of the physiological significance of plant endopolyploidy has remained elusive. Here, we modeled and experimentally verified a high-resolution DNA endoploidy map of the developing Arabidopsis thaliana root, revealing a remarkable spatiotemporal control of DNA endoploidy levels across tissues. Fitting of a simplified model to publicly available data sets profiling root gene expression under various environmental stress conditions suggested that this root endoploidy patterning may be stress-responsive. Furthermore, cellular and transcriptomic analyses revealed that inhibition of endoreplication onset alters the nuclear-to-cellular volume ratio and the expression of cell wall-modifying genes, in correlation with the appearance of cell structural changes. Our data indicate that endopolyploidy might serve to coordinate cell expansion with structural stability and that spatiotemporal endoreplication pattern changes may buffer for stress conditions, which may explain the widespread occurrence of the endocycle in plant species growing in extreme or variable environments.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures







References
-
- Artlip T.S., Madison J.T., Setter T.L. (1995). Water deficit in developing endosperm of maize: cell division and nuclear DNA endoreduplication. Plant Cell Environ. 18: 1034–1040.
-
- Barow M. (2006). Endopolyploidy in seed plants. BioEssays 28: 271–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

