Size control in mammalian cells involves modulation of both growth rate and cell cycle duration
- PMID: 30115907
- PMCID: PMC6095894
- DOI: 10.1038/s41467-018-05393-0
Size control in mammalian cells involves modulation of both growth rate and cell cycle duration
Abstract
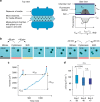
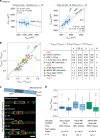
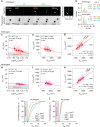
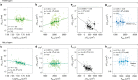
Despite decades of research, how mammalian cell size is controlled remains unclear because of the difficulty of directly measuring growth at the single-cell level. Here we report direct measurements of single-cell volumes over entire cell cycles on various mammalian cell lines and primary human cells. We find that, in a majority of cell types, the volume added across the cell cycle shows little or no correlation to cell birth size, a homeostatic behavior called "adder". This behavior involves modulation of G1 or S-G2 duration and modulation of growth rate. The precise combination of these mechanisms depends on the cell type and the growth condition. We have developed a mathematical framework to compare size homeostasis in datasets ranging from bacteria to mammalian cells. This reveals that a near-adder behavior is the most common type of size control and highlights the importance of growth rate modulation to size control in mammalian cells.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

