Structural Implications of Genotypic Variations in HIV-1 Integrase From Diverse Subtypes
- PMID: 30116231
- PMCID: PMC6083056
- DOI: 10.3389/fmicb.2018.01754
Structural Implications of Genotypic Variations in HIV-1 Integrase From Diverse Subtypes
Abstract
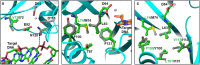
Human immunodeficiency virus type 1 (HIV-1) integrase (IN) integrates viral DNA into the host genome using its 3'-end processing and strand-transfer activities. Due to the importance of HIV-1 IN, it is targeted by the newest class of approved drugs known as integrase strand transfer inhibitors (INSTIs). INSTIs are efficient in maintaining low viral load; however, as with other approved antivirals, resistance mutations emerge in patients receiving INSTI-containing therapy. As INSTIs are becoming increasingly accessible worldwide, it is important to understand the mechanism(s) of INSTI susceptibility. There is strong evidence suggesting differences in the patterns and mechanisms of drug resistance between HIV-1 subtype B, which dominates in United States, Western Europe and Australia, and non-B infections that are most prevalent in countries of Africa and Asia. IN polymorphisms and other genetic differences among diverse subtypes are likely responsible for these different patterns, but lack of a full-length high-resolution structure of HIV-1 IN has been a roadblock in understanding the molecular mechanisms of INSTI resistance and the impact of polymorphisms on therapy outcome. A recently reported full-length medium-resolution cryoEM structure of HIV-1 IN provides insights into understanding the mechanism of integrase function and the impact of genetic variation on the effectiveness of INSTIs. Here we use molecular modeling to explore the structural impact of IN polymorphisms on the IN reaction mechanism and INSTI susceptibility.
Keywords: HIV-1 integrase; HIV-1 subtypes; drug-resistance; polymorphism; strand transfer inhibitor.
Figures




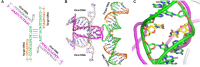
References
-
- Brado D., Obasa A. E., Ikomey G. M., Cloete R., Singh K., Engelbrecht S., et al. (2018). Analyses of HIV-1 integrase sequences prior to South African national HIV-treatment program and available of integrase inhibitors in Cape Town, South Africa. Sci. Rep. 8 4709–4718. 10.1038/s41598-018-22914-5 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

