A Simple Separation Method of the Protein and Polystyrene Bead-Labeled Protein for Enhancing the Performance of Fluorescent Sensor
- PMID: 30116650
- PMCID: PMC6079413
- DOI: 10.1155/2018/8461380
A Simple Separation Method of the Protein and Polystyrene Bead-Labeled Protein for Enhancing the Performance of Fluorescent Sensor
Abstract
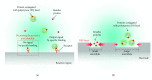
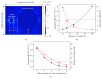
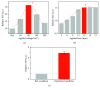
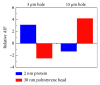
Dielectrophoresis- (DEP-) based separation method between a protein, amyloid beta 42, and polystyrene (PS) beads in different microholes was demonstrated for enhancement of performance for bead-based fluorescent sensor. An intensity of ∇|E|2 was relative to a diameter of a microhole, and the diameters of two microholes for separation between the protein and PS beads were simulated to 3 μm and 15 μm, respectively. The microholes were fabricated by microelectromechanical systems (MEMS). The separation between the protein and the PS beads was demonstrated by comparing the average intensity of fluorescence (AIF) by each molecule. Relative AIF was measured in various applying voltage and time conditions, and the conditions for allocating the PS beads into 15 μm hole were optimized at 80 mV and 15 min, respectively. In the optimized condition, the relative AIF was observed approximately 4.908 ± 0.299. Finally, in 3 μm and 15 μm hole, the AIFs were approximately 3.143 and -1.346 by 2 nm of protein and about -2.515 and 4.211 by 30 nm of the PS beads, respectively. The results showed that 2 nm of the protein and 30 nm of PS beads were separated by DEP force in each microhole effectively, and that our method is applicable as a new method to verify an efficiency of the labeling for bead-based fluorescent sensor ∇|E|2.
Figures

Similar articles
-
A review of polystyrene bead manipulation by dielectrophoresis.RSC Adv. 2019 Feb 8;9(9):4963-4981. doi: 10.1039/c8ra09017c. eCollection 2019 Feb 5. RSC Adv. 2019. PMID: 35514668 Free PMC article. Review.
-
Erratum: Preparation of Poly(pentafluorophenyl acrylate) Functionalized SiO2 Beads for Protein Purification.J Vis Exp. 2019 Apr 30;(146). doi: 10.3791/6328. J Vis Exp. 2019. PMID: 31038480
-
Selective nanomanipulation of fluorescent polystyrene nano-beads and single quantum dots at gold nanostructures based on the AC-dielectrophoretic force.Nanoscale. 2015 Dec 21;7(47):20277-83. doi: 10.1039/c5nr05963a. Epub 2015 Nov 18. Nanoscale. 2015. PMID: 26579981
-
A low sample volume particle separation device with electrokinetic pumping based on circular travelling-wave electroosmosis.Lab Chip. 2013 Aug 7;13(15):3082-9. doi: 10.1039/c3lc50343g. Lab Chip. 2013. PMID: 23753015
-
Dielectrophoresis Manipulation: Versatile Lateral and Vertical Mechanisms.Biosensors (Basel). 2019 Feb 26;9(1):30. doi: 10.3390/bios9010030. Biosensors (Basel). 2019. PMID: 30813614 Free PMC article. Review.
References
-
- Sahoo H. Fluorescent labeling techniques in biomolecules: a flashback. RSC Advances. 2012;2(18):7017–7029. doi: 10.1039/c2ra20389h. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

