A patient-derived explant (PDE) model of hormone-dependent cancer
- PMID: 30117261
- PMCID: PMC6120230
- DOI: 10.1002/1878-0261.12354
A patient-derived explant (PDE) model of hormone-dependent cancer
Abstract
Breast and prostate cancer research to date has largely been predicated on the use of cell lines in vitro or in vivo. These limitations have led to the development of more clinically relevant models, such as organoids or murine xenografts that utilize patient-derived material; however, issues related to low take rate, long duration of establishment, and the associated costs constrain use of these models. This study demonstrates that ex vivo culture of freshly resected breast and prostate tumor specimens obtained from surgery, termed patient-derived explants (PDEs), provides a high-throughput and cost-effective model that retains the native tissue architecture, microenvironment, cell viability, and key oncogenic drivers. The PDE model provides a unique approach for direct evaluation of drug responses on an individual patient's tumor, which is amenable to analysis using contemporary genomic technologies. The ability to rapidly evaluate drug efficacy in patient-derived material has high potential to facilitate implementation of personalized medicine approaches.
Keywords: ex vivo culture; patient-derived explant; preclinical tumor model.
© 2018 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures


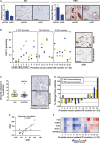
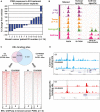
References
-
- Ali S and Coombes RC (2002) Endocrine‐responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2, 101–112. - PubMed
-
- Arnedos M, Vielh P, Soria JC, Andre F (2013) The genetic complexity of common cancers and the promise of personalized medicine: is there any hope? J Pathol 232, 274–282. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

