An Arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton
- PMID: 30117653
- PMCID: PMC6381789
- DOI: 10.1111/pbi.12998
An Arabidopsis E3 ligase HUB2 increases histone H2B monoubiquitination and enhances drought tolerance in transgenic cotton
Abstract
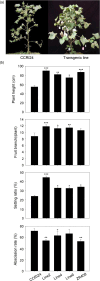
The HUB2 gene encoding histone H2B monoubiquitination E3 ligase is involved in seed dormancy, flowering timing, defence response and salt stress regulation in Arabidopsis thaliana. In this study, we used the cauliflower mosaic virus (CaMV) 35S promoter to drive AtHUB2 overexpression in cotton and found that it can significantly improve the agricultural traits of transgenic cotton plants under drought stress conditions, including increasing the fruit branch number, boll number, and boll-setting rate and decreasing the boll abscission rate. In addition, survival and soluble sugar, proline and leaf relative water contents were increased in transgenic cotton plants after drought stress treatment. In contrast, RNAi knockdown of GhHUB2 genes reduced the drought resistance of transgenic cotton plants. AtHUB2 overexpression increased the global H2B monoubiquitination (H2Bub1) level through a direct interaction with GhH2B1 and up-regulated the expression of drought-related genes in transgenic cotton plants. Furthermore, we found a significant increase in H3K4me3 at the DREB locus in transgenic cotton, although no change in H3K4me3 was identified at the global level. These results demonstrated that AtHUB2 overexpression changed H2Bub1 and H3K4me3 levels at the GhDREB chromatin locus, leading the GhDREB gene to respond quickly to drought stress to improve transgenic cotton drought resistance, but had no influence on transgenic cotton development under normal growth conditions. Our findings also provide a useful route for breeding drought-resistant transgenic plants.
Keywords: HISTONE MONOUBIQUITINATION 2 (HUB2); Gossypium hirsutum Linn.; drought; histone methylation; histone monoubiquitination; transgenic plants.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Agarwal, P. , Agarwal, P.K. , Nair, S. , Sopory, S.K. and Reddy, M.K. (2007) Stress‐inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA‐binding activity. Mol. Genet. Genomics, 277, 189–198. - PubMed
-
- Bartels, D. and Sunkar, R. (2005) Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58.
-
- Bates, L.S. , Waldren, R.P. and Teare, I.D. (1973) Rapid determination of free proline for water‐stress studies. Plant Soil, 39, 205–207.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

