Improved Sensitivity and Separations for Phosphopeptides using Online Liquid Chromotography Coupled with Structures for Lossless Ion Manipulations Ion Mobility-Mass Spectrometry
- PMID: 30118596
- PMCID: PMC6211290
- DOI: 10.1021/acs.analchem.8b02397
Improved Sensitivity and Separations for Phosphopeptides using Online Liquid Chromotography Coupled with Structures for Lossless Ion Manipulations Ion Mobility-Mass Spectrometry
Abstract
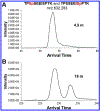
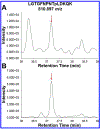
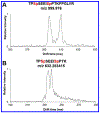
Phosphoproteomics greatly augments proteomics and holds tremendous potential for insights into the modulation of biological systems for various disease states. However, numerous challenges hinder conventional methods in terms of measurement sensitivity, throughput, quantification, and capabilities for confident phosphopeptide and phosphosite identification. In this work, we report the first example of integrating structures for lossless ion manipulations ion mobility-mass spectrometry (SLIM IM-MS) with online reversed-phase liquid chromatography (LC) to evaluate its potential for addressing the aforementioned challenges. A mixture of 51 heavy-labeled phosphopeptides was analyzed with a SLIM IM module having integrated ion accumulation and long-path separation regions. The SLIM IM-MS provided limits of detection as low as 50-100 pM (50-100 amol/μL) for several phosphopeptides, with the potential for significant further improvements. In addition, conventionally problematic phosphopeptide isomers could be resolved following an 18 m SLIM IM separation. The 2-D LC-IM peak capacity was estimated as ∼9000 for a 90 min LC separation coupled to an 18 m SLIM IM separation, considerably higher than LC alone and providing a basis for both improved identification and quantification, with additional gains projected with the future use of longer path SLIM IM separations. Thus, LC-SLIM IM-MS offers great potential for improving the sensitivity, separation, and throughput of phosphoproteomics analyses.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

