An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses
- PMID: 30118682
- PMCID: PMC6329589
- DOI: 10.1016/j.molcel.2018.07.010
An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses
Abstract
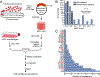
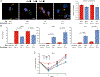
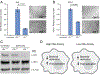
Although macrophages are armed with potent antibacterial functions, Mycobacterium tuberculosis (Mtb) replicates inside these innate immune cells. Determinants of macrophage intrinsic bacterial control, and the Mtb strategies to overcome them, are poorly understood. To further study these processes, we used an affinity tag purification mass spectrometry (AP-MS) approach to identify 187 Mtb-human protein-protein interactions (PPIs) involving 34 secreted Mtb proteins. This interaction map revealed two factors involved in Mtb pathogenesis-the secreted Mtb protein, LpqN, and its binding partner, the human ubiquitin ligase CBL. We discovered that an lpqN Mtb mutant is attenuated in macrophages, but growth is restored when CBL is removed. Conversely, Cbl-/- macrophages are resistant to viral infection, indicating that CBL regulates cell-intrinsic polarization between antibacterial and antiviral immunity. Collectively, these findings illustrate the utility of this Mtb-human PPI map for developing a deeper understanding of the intricate interactions between Mtb and its host.
Keywords: Cbl; LpqN; host-pathogen interaction; macrophage; mycobacterium; protein-protein interaction; tuberculosis; ubiquitin.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTEREST
Daniel A. Portnoy has a financial interest in Aduro Biotech, and both he and the company stand to benefit from commercialization of this research.
Figures



Comment in
-
Bacterial Protein Reshapes Host Defense toward Antiviral Responses.Mol Cell. 2018 Aug 16;71(4):483-484. doi: 10.1016/j.molcel.2018.08.006. Mol Cell. 2018. PMID: 30118676 Free PMC article.
References
-
- Alberts B (1998). The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 92, 291–294. - PubMed
-
- Bach H, Papavinasasundaram KG, Wong D, Hmama Z, and Av-Gay Y (2008). Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host & Microbe 3, 316–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

