Functional CRISPR screen identifies AP1-associated enhancer regulating FOXF1 to modulate oncogene-induced senescence
- PMID: 30119690
- PMCID: PMC6097335
- DOI: 10.1186/s13059-018-1494-1
Functional CRISPR screen identifies AP1-associated enhancer regulating FOXF1 to modulate oncogene-induced senescence
Abstract
Background: Functional characterization of non-coding elements in the human genome is a major genomic challenge and the maturation of genome-editing technologies is revolutionizing our ability to achieve this task. Oncogene-induced senescence, a cellular state of irreversible proliferation arrest that is enforced following excessive oncogenic activity, is a major barrier against cancer transformation; therefore, bypassing oncogene-induced senescence is a critical step in tumorigenesis. Here, we aim at further identification of enhancer elements that are required for the establishment of this state.
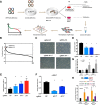
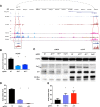
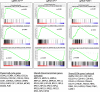
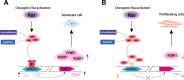
Results: We first apply genome-wide profiling of enhancer-RNAs (eRNAs) to systematically identify enhancers that are activated upon oncogenic stress. DNA motif analysis of these enhancers indicates AP-1 as a major regulator of the transcriptional program induced by oncogene-induced senescence. We thus constructed a CRISPR-Cas9 sgRNA library designed to target senescence-induced enhancers that are putatively regulated by AP-1 and used it in a functional screen. We identify a critical enhancer that we name EnhAP1-OIS1 and validate that mutating the AP-1 binding site within this element results in oncogene-induced senescence bypass. Furthermore, we identify FOXF1 as the gene regulated by this enhancer and demonstrate that FOXF1 mediates EnhAP1-OIS1 effect on the senescence phenotype.
Conclusions: Our study elucidates a novel cascade mediated by AP-1 and FOXF1 that regulates oncogene-induced senescence and further demonstrates the power of CRISPR-based functional genomic screens in deciphering the function of non-coding regulatory elements in the genome.
Keywords: AP1; CRISPR; Enhancers; FOS; FOXF1; Functional screen; Gene regulation; JUN; Oncogene-induced senescence.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

