Biochemical properties and in planta effects of NopM, a rhizobial E3 ubiquitin ligase
- PMID: 30120198
- PMCID: PMC6166720
- DOI: 10.1074/jbc.RA118.004444
Biochemical properties and in planta effects of NopM, a rhizobial E3 ubiquitin ligase
Abstract
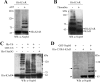
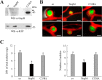
Nodulation outer protein M (NopM) is an IpaH family type three (T3) effector secreted by the nitrogen-fixing nodule bacterium Sinorhizobium sp. strain NGR234. Previous work indicated that NopM is an E3 ubiquitin ligase required for an optimal symbiosis between NGR234 and the host legume Lablab purpureus Here, we continued to analyze the function of NopM. Recombinant NopM was biochemically characterized using an in vitro ubiquitination system with Arabidopsis thaliana proteins. In this assay, NopM forms unanchored polyubiquitin chains and possesses auto-ubiquitination activity. In a NopM variant lacking any lysine residues, auto-ubiquitination was not completely abolished, indicating noncanonical auto-ubiquitination of the protein. In addition, we could show intermolecular ubiquitin transfer from NopM to C338A (enzymatically inactive NopM form) in vitro Bimolecular fluorescence complementation analysis provided clues about NopM-NopM interactions at plasma membranes in planta NopM, but not C338A, expressed in tobacco cells induced cell death, suggesting that E3 ubiquitin ligase activity of NopM induced effector-triggered immunity responses. Likewise, expression of NopM in Lotus japonicus caused reduced nodule formation, whereas expression of C338A showed no obvious effects on symbiosis. Further experiments indicated that serine residue 26 of NopM is phosphorylated in planta and that NopM can be phosphorylated in vitro by salicylic acid-induced protein kinase (NtSIPK), a mitogen-activated protein kinase (MAPK) of tobacco. Hence, NopM is a phosphorylated T3 effector that can interact with itself, with ubiquitin, and with MAPKs.
Keywords: E3 ubiquitin ligase; bacteria; effector protein; phosphorylation; symbiosis; type III secretion system (T3SS); ubiquitination.
© 2018 Xu et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

