Tdp-25 Routing to Autophagy and Proteasome Ameliorates its Aggregation in Amyotrophic Lateral Sclerosis Target Cells
- PMID: 30120266
- PMCID: PMC6098007
- DOI: 10.1038/s41598-018-29658-2
Tdp-25 Routing to Autophagy and Proteasome Ameliorates its Aggregation in Amyotrophic Lateral Sclerosis Target Cells
Abstract
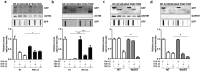
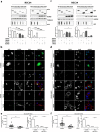
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that primarily affects motoneurons, while non-neuronal cells may contribute to disease onset and progression. Most ALS cases are characterized by the mislocalization and aggregation of the TAR DNA binding protein 43 (TDP-43) in affected cells. TDP-43 aggregates contain C-terminal TDP-43 fragments of 35 kDa (TDP-35) and 25 kDa (TDP-25) and have been mainly studied in motoneurons, while little is currently known about their rate of accumulation and clearance in myoblasts. Here, we performed a comparative study in immortalized motoneuronal like (NSC34; i-motoneurons) cells and stabilized myoblasts (C2C12; s-myoblasts) to evaluate if these two cell types differentially accumulate and clear TDP forms. The most aggregating specie in i-motoneurons is the TDP-25 fragment, mainly constituted by the "prion-like" domain of TDP-43. To a lower extent, TDP-25 also aggregates in s-myoblasts. In both cell types, all TDP species are cleared by proteasome, but TDP-25 impairs autophagy. Interestingly, the routing of TDP-25 fragment to proteasome, by overexpressing BAG1, or to autophagy, by overexpressing HSPB8 or BAG3 decreased its accumulation in both cell types. These results demonstrate that promoting the chaperone-assisted clearance of ALS-linked proteins is beneficial not only in motoneurons but also in myoblasts.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Comm351, 602–611, 10.1016/j.bbrc.2006.10.093 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- InEurope short stay grant/International Brain Research Organization (IBRO)/International
- 537 - 2015/European Molecular Biology Organization (EMBO)/International
- FFABR (Fondo per il Finanziamento delle Attività Base di Ricerca)/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- FFABR (Fondo per il Finanziamento delle Attività Base di Ricerca)/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- Fondo per il Finanziamento delle Attività Base di Ricerca (FFABR)/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- 2015LFPNMN/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- Fondo per il Finanziamento delle Attività Base di Ricerca (FFABR)/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- 2015LFPNMN/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)/International
- 2014-0686/Fondazione Cariplo (Cariplo Foundation)/International
- 2017-0000/Fondazione Cariplo (Cariplo Foundation)/International
- 2014-0686/Fondazione Cariplo (Cariplo Foundation)/International
- Co_ALS/Ministry of Health, Italy | Agenzia Italiana del Farmaco, Ministero della Salute (Italian Medicines Agency)/International
- Co_ALS/Ministry of Health, Italy | Agenzia Italiana del Farmaco, Ministero della Salute (Italian Medicines Agency)/International
- GR-2011-0234/Ministero della Salute (Ministry of Health, Italy)/International
- GGP14039/Fondazione Telethon (Telethon Foundation)/International
- 16406/AFM-Téléthon (French Muscular Dystrophy Association)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

