QM/MM Description of Newly Selected Catalytic Bioscavengers Against Organophosphorus Compounds Revealed Reactivation Stimulus Mediated by Histidine Residue in the Acyl-Binding Loop
- PMID: 30123127
- PMCID: PMC6085465
- DOI: 10.3389/fphar.2018.00834
QM/MM Description of Newly Selected Catalytic Bioscavengers Against Organophosphorus Compounds Revealed Reactivation Stimulus Mediated by Histidine Residue in the Acyl-Binding Loop
Abstract
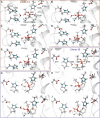
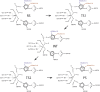
Butyrylcholinesterase (BChE) is considered as an efficient stoichiometric antidote against organophosphorus (OP) poisons. Recently we utilized combination of calculations and ultrahigh-throughput screening (uHTS) to select BChE variants capable of catalytic destruction of OP pesticide paraoxon. The purpose of this study was to elucidate the molecular mechanism underlying enzymatic hydrolysis of paraoxon by BChE variants using hybrid quantum mechanical/molecular mechanical (QM/MM) calculations. Detailed analysis of accomplished QM/MM runs revealed that histidine residues introduced into the acyl-binding loop are always located in close proximity with aspartate residue at position 70. Histidine residue acts as general base thus leading to attacking water molecule activation and subsequent SN2 inline hydrolysis resulting in BChE reactivation. This combination resembles canonical catalytic triad found in active centers of various proteases. Carboxyl group activates histidine residue by altering its pKa, which in turn promotes the activation of water molecule in terms of its nucleophilicity. Observed re-protonation of catalytic serine residue at position 198 from histidine residue at position 438 recovers initial configuration of the enzyme's active center, facilitating next catalytic cycle. We therefore suggest that utilization of uHTS platform in combination with deciphering of molecular mechanisms by QM/MM calculations may significantly improve our knowledge of enzyme function, propose new strategies for enzyme design and open new horizons in generation of catalytic bioscavengers against OP poisons.
Keywords: bioscavenger; butyrylcholinesterase; computer design; organophosphorus compound; paraoxon; ultrahigh-throughput screening.
Figures



References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Adamo C., Barone V. (1998). Toward chemical accuracy in the computation of NMR shieldings: the PBE0 model. Chem. Phys. Lett. 298 113–119. 10.1016/S0009-2614(98)01201-9 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

