PU.1 regulates Alzheimer's disease-associated genes in primary human microglia
- PMID: 30124174
- PMCID: PMC6102813
- DOI: 10.1186/s13024-018-0277-1
PU.1 regulates Alzheimer's disease-associated genes in primary human microglia
Abstract
Background: Microglia play critical roles in the brain during homeostasis and pathological conditions. Understanding the molecular events underpinning microglial functions and activation states will further enable us to target these cells for the treatment of neurological disorders. The transcription factor PU.1 is critical in the development of myeloid cells and a major regulator of microglial gene expression. In the brain, PU.1 is specifically expressed in microglia and recent evidence from genome-wide association studies suggests that reductions in PU.1 contribute to a delayed onset of Alzheimer's disease (AD), possibly through limiting neuroinflammatory responses.
Methods: To investigate how PU.1 contributes to immune activation in human microglia, microarray analysis was performed on primary human mixed glial cultures subjected to siRNA-mediated knockdown of PU.1. Microarray hits were confirmed by qRT-PCR and immunocytochemistry in both mixed glial cultures and isolated microglia following PU.1 knockdown. To identify attenuators of PU.1 expression in microglia, high throughput drug screening was undertaken using a compound library containing FDA-approved drugs. NanoString and immunohistochemistry was utilised to investigate the expression of PU.1 itself and PU.1-regulated mediators in primary human brain tissue derived from neurologically normal and clinically and pathologically confirmed cases of AD.
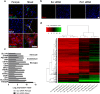
Results: Bioinformatic analysis of gene expression upon PU.1 silencing in mixed glial cultures revealed a network of modified AD-associated microglial genes involved in the innate and adaptive immune systems, particularly those involved in antigen presentation and phagocytosis. These gene changes were confirmed using isolated microglial cultures. Utilising high throughput screening of FDA-approved compounds in mixed glial cultures we identified the histone deacetylase inhibitor vorinostat as an effective attenuator of PU.1 expression in human microglia. Further characterisation of vorinostat in isolated microglial cultures revealed gene and protein changes partially recapitulating those seen following siRNA-mediated PU.1 knockdown. Lastly, we demonstrate that several of these PU.1-regulated genes are expressed by microglia in the human AD brain in situ.
Conclusions: Collectively, these results suggest that attenuating PU.1 may be a valid therapeutic approach to limit microglial-mediated inflammatory responses in AD and demonstrate utility of vorinostat for this purpose.
Keywords: Alzheimer’s disease; Antigen presentation; Drug screening; Neuroinflammation; Phagocytosis; Vorinostat.
Conflict of interest statement
Ethics approval and consent to participate
All brain tissue collection and processing protocols were approved by the Northern Regional Ethics Committee (New Zealand) for biopsy tissue, and the University of Auckland Human Participants Ethics Committee (New Zealand) for the post-mortem brain tissue. All methods were carried out in accordance with the approved guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

