Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine
- PMID: 30124358
- PMCID: PMC6207407
- DOI: 10.1089/omi.2018.0097
Rise of Deep Learning for Genomic, Proteomic, and Metabolomic Data Integration in Precision Medicine
Abstract
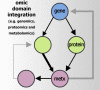
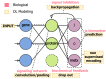
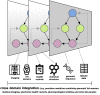
Machine learning (ML) is being ubiquitously incorporated into everyday products such as Internet search, email spam filters, product recommendations, image classification, and speech recognition. New approaches for highly integrated manufacturing and automation such as the Industry 4.0 and the Internet of things are also converging with ML methodologies. Many approaches incorporate complex artificial neural network architectures and are collectively referred to as deep learning (DL) applications. These methods have been shown capable of representing and learning predictable relationships in many diverse forms of data and hold promise for transforming the future of omics research and applications in precision medicine. Omics and electronic health record data pose considerable challenges for DL. This is due to many factors such as low signal to noise, analytical variance, and complex data integration requirements. However, DL models have already been shown capable of both improving the ease of data encoding and predictive model performance over alternative approaches. It may not be surprising that concepts encountered in DL share similarities with those observed in biological message relay systems such as gene, protein, and metabolite networks. This expert review examines the challenges and opportunities for DL at a systems and biological scale for a precision medicine readership.
Keywords: artificial intelligence; biomarkers; deep learning; machine learning; multiomics data integration; precision medicine.
Conflict of interest statement
D.G. is the Director of Data Science and Bioinformatics at CDS—Creative Data Solutions LLC,
Figures
References
-
- Alipanahi B, Delong A, Weirauch MT, and Frey BJ. (2015). Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33, 831–838 - PubMed
-
- Breiman L. (2001). Random forests. Machine Learning 45, 5–32
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
