i-Motif DNA: structural features and significance to cell biology
- PMID: 30124962
- PMCID: PMC6144788
- DOI: 10.1093/nar/gky735
i-Motif DNA: structural features and significance to cell biology
Abstract
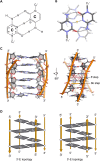
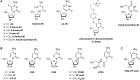
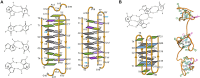
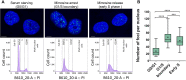
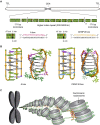
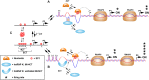
The i-motif represents a paradigmatic example of the wide structural versatility of nucleic acids. In remarkable contrast to duplex DNA, i-motifs are four-stranded DNA structures held together by hemi- protonated and intercalated cytosine base pairs (C:C+). First observed 25 years ago, and considered by many as a mere structural oddity, interest in and discussion on the biological role of i-motifs have grown dramatically in recent years. In this review we focus on structural aspects of i-motif formation, the factors leading to its stabilization and recent studies describing the possible role of i-motifs in fundamental biological processes.
Figures

References
-
- Watson J.D., Crick F.H.C.. Molecular structure of nucleic acids - a structure for deoxyribose nucleic acid. Nature. 1953; 171:737–738. - PubMed
-
- Du Y., Zhou X.. Targeting non-B-form DNA in living cells. Chem. Rec. 2013; 13:371–384. - PubMed
-
- Choi J., Majima T.. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011; 40:5893–5909. - PubMed
-
- Sen D., Gilbert W.. Formation of parallel 4-Stranded complexes by Guanine-Rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. - PubMed
-
- Gehring K., Leroy J.L., Gueron M.. A tetrameric DNA-structure with protonated cytosine.cytosine Base-Pairs. Nature. 1993; 363:561–565. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

