Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10
- PMID: 30125002
- PMCID: PMC6212717
- DOI: 10.1093/nar/gky737
Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10
Abstract
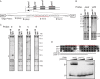
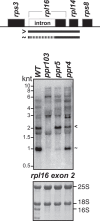
Pentatricopeptide repeat (PPR) proteins are a large family of helical repeat proteins that bind RNA in mitochondria and chloroplasts. Sites of PPR action have been inferred primarily from genetic data, which have led to the view that most PPR proteins act at a very small number of sites in vivo. Here, we report new functions for three chloroplast PPR proteins that had already been studied in depth. Maize PPR5, previously shown to promote trnG splicing, is also required for rpl16 splicing. Maize PPR10, previously shown to bind the atpI-atpH and psaJ-rpl33 intercistronic regions, also stabilizes a 3'-end downstream from psaI. Arabidopsis PGR3, shown previously to bind upstream of petL, also binds the rpl14-rps8 intercistronic region where it stabilizes a 3'-end and stimulates rps8 translation. These functions of PGR3 are conserved in maize. The discovery of new functions for three proteins that were already among the best characterized members of the PPR family implies that functional repertoires of PPR proteins are more complex than have been appreciated. The diversity of sequences bound by PPR10 and PGR3 in vivo highlights challenges of predicting binding sites of native PPR proteins based on the amino acid code for nucleotide recognition by PPR motifs.
Figures



Similar articles
-
RNA-binding specificity landscape of the pentatricopeptide repeat protein PPR10.RNA. 2017 Apr;23(4):586-599. doi: 10.1261/rna.059568.116. Epub 2017 Jan 20. RNA. 2017. PMID: 28108520 Free PMC article.
-
The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts.Mol Cell Biol. 2008 Sep;28(17):5337-47. doi: 10.1128/MCB.00563-08. Epub 2008 Jun 30. Mol Cell Biol. 2008. PMID: 18591259 Free PMC article.
-
Structural basis for the modular recognition of single-stranded RNA by PPR proteins.Nature. 2013 Dec 5;504(7478):168-71. doi: 10.1038/nature12651. Epub 2013 Oct 27. Nature. 2013. PMID: 24162847
-
PPR-SMRs: ancient proteins with enigmatic functions.RNA Biol. 2013;10(9):1501-10. doi: 10.4161/rna.26172. Epub 2013 Aug 28. RNA Biol. 2013. PMID: 24004908 Free PMC article. Review.
-
Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants.Plant Cell Physiol. 2012 Jul;53(7):1171-9. doi: 10.1093/pcp/pcs069. Epub 2012 May 9. Plant Cell Physiol. 2012. PMID: 22576772 Review.
Cited by
-
The Chloroplast RNA Binding Protein CP31A Has a Preference for mRNAs Encoding the Subunits of the Chloroplast NAD(P)H Dehydrogenase Complex and Is Required for Their Accumulation.Int J Mol Sci. 2020 Aug 6;21(16):5633. doi: 10.3390/ijms21165633. Int J Mol Sci. 2020. PMID: 32781615 Free PMC article.
-
Identifying Plant Pentatricopeptide Repeat Coding Gene/Protein Using Mixed Feature Extraction Methods.Front Plant Sci. 2019 Jan 10;9:1961. doi: 10.3389/fpls.2018.01961. eCollection 2018. Front Plant Sci. 2019. PMID: 30687359 Free PMC article.
-
An RNA Chaperone-Like Protein Plays Critical Roles in Chloroplast mRNA Stability and Translation in Arabidopsis and Maize.Plant Cell. 2019 Jun;31(6):1308-1327. doi: 10.1105/tpc.18.00946. Epub 2019 Apr 8. Plant Cell. 2019. PMID: 30962391 Free PMC article.
-
Expanding the repertoire of human tandem repeat RNA-binding proteins.PLoS One. 2023 Sep 20;18(9):e0290890. doi: 10.1371/journal.pone.0290890. eCollection 2023. PLoS One. 2023. PMID: 37729217 Free PMC article.
-
Plastid retrograde signaling: A developmental perspective.Plant Cell. 2024 Oct 3;36(10):3903-3913. doi: 10.1093/plcell/koae094. Plant Cell. 2024. PMID: 38546347 Free PMC article. Review.
References
-
- Bock R. Bock R. Structure, function, and inheritance of plastid genomes. Cell and Molecular Biology of Plastids. 2007; Heidelberg: Springer-Verlag; 29–63.
-
- Borner T., Aleynikova A.Y., Zubo Y.O., Kusnetsov V.V.. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta. 2015; 1847:761–769. - PubMed
-
- Germain A., Hotto A.M., Barkan A., Stern D.B.. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA. 2013; 4:295–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

