Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae
- PMID: 30125091
- PMCID: PMC6145841
- DOI: 10.1021/acs.biochem.8b00653
Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae
Abstract
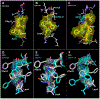
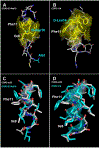
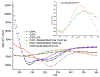
Streptococcus pneumoniae is an important pathogen that utilizes quorum sensing (QS) to regulate genetic transformation, virulence, and biofilm formation. The competence-stimulating peptide (CSP) is a 17-amino acid signal peptide that is used by S. pneumoniae to trigger QS. S. pneumoniae strains can be divided into two main specificity groups based on the CSP signal they produce (CSP1 or CSP2) and their compatible receptors (ComD1 or ComD2, respectively). Modulation of QS in S. pneumoniae can be achieved by targeting the CSP:ComD interaction using synthetic CSP analogues. However, to rationally design CSP-based QS modulators with enhanced activities, an in-depth understanding of the structural features that are required for receptor binding is needed. Herein, we report a comprehensive in-solution three-dimensional structural characterization of eight CSP1 and CSP2 analogues with varied biological activities using nuclear magnetic resonance spectroscopy. Analysis of these structures revealed two distinct hydrophobic patches required for effective ComD1 and ComD2 binding.
Figures



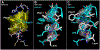

References
-
- Mehr S, and Wood N (2012) Streptococcus pneumoniae--A Review of Carriage, Infection, Serotype Replacement and Vaccination, Paediatr Respir Rev 13, 258–264. - PubMed
-
- Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, Moore MR, Zell ER, Linder JA, Grijalva CG, Metlay JP, and Finkelstein JA (2011) Healthcare Utilization and Cost of Pneumococcal Disease in the United States, Vaccine 29, 3398–3412. - PubMed
-
- Linares J, Ardanuy C, Pallares R, and Fenoll A (2010) Changes in Antimicrobial Resistance, Serotypes and Genotypes in Streptococcus pneumoniae Over a 30-Year Period, Clin Microbiol Infect 16, 402–410 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

