Timescales of influenza A/H3N2 antibody dynamics
- PMID: 30125272
- PMCID: PMC6117086
- DOI: 10.1371/journal.pbio.2004974
Timescales of influenza A/H3N2 antibody dynamics
Abstract
Human immunity influences the evolution and impact of influenza strains. Because individuals are infected with multiple influenza strains during their lifetime, and each virus can generate a cross-reactive antibody response, it is challenging to quantify the processes that shape observed immune responses or to reliably detect recent infection from serological samples. Using a Bayesian model of antibody dynamics at multiple timescales, we explain complex cross-reactive antibody landscapes by inferring participants' histories of infection with serological data from cross-sectional and longitudinal studies of influenza A/H3N2 in southern China and Vietnam. We find that individual-level influenza antibody profiles can be explained by a short-lived, broadly cross-reactive response that decays within a year to leave a smaller long-term response acting against a narrower range of strains. We also demonstrate that accounting for dynamic immune responses alongside infection history can provide a more accurate alternative to traditional definitions of seroconversion for the estimation of infection attack rates. Our work provides a general model for quantifying aspects of influenza immunity acting at multiple timescales based on contemporary serological data and suggests a two-armed immune response to influenza infection consistent with competitive dynamics between B cell populations. This approach to analysing multiple timescales for antigenic responses could also be applied to other multistrain pathogens such as dengue and related flaviviruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
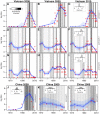
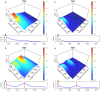

References
-
- Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. The Lancet. 2010;375(1100–1108). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

