An EBNA3C-deleted Epstein-Barr virus (EBV) mutant causes B-cell lymphomas with delayed onset in a cord blood-humanized mouse model
- PMID: 30125329
- PMCID: PMC6117096
- DOI: 10.1371/journal.ppat.1007221
An EBNA3C-deleted Epstein-Barr virus (EBV) mutant causes B-cell lymphomas with delayed onset in a cord blood-humanized mouse model
Abstract
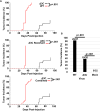
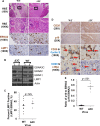
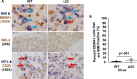
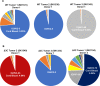
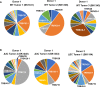
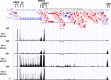
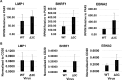
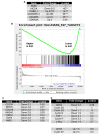
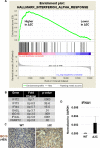
EBV causes human B-cell lymphomas and transforms B cells in vitro. EBNA3C, an EBV protein expressed in latently-infected cells, is required for EBV transformation of B cells in vitro. While EBNA3C undoubtedly plays a key role in allowing EBV to successfully infect B cells, many EBV+ lymphomas do not express this protein, suggesting that cellular mutations and/or signaling pathways may obviate the need for EBNA3C in vivo under certain conditions. EBNA3C collaborates with EBNA3A to repress expression of the CDKN2A-encoded tumor suppressors, p16 and p14, and EBNA3C-deleted EBV transforms B cells containing a p16 germline mutation in vitro. Here we have examined the phenotype of an EBNAC-deleted virus (Δ3C EBV) in a cord blood-humanized mouse model (CBH). We found that the Δ3C virus induced fewer lymphomas (occurring with a delayed onset) in comparison to the wild-type (WT) control virus, although a subset (10/26) of Δ3C-infected CBH mice eventually developed invasive diffuse large B cell lymphomas with type III latency. Both WT and Δ3C viruses induced B-cell lymphomas with restricted B-cell populations and heterogeneous T-cell infiltration. In comparison to WT-infected tumors, Δ3C-infected tumors had greatly increased p16 levels, and RNA-seq analysis revealed a decrease in E2F target gene expression. However, we found that Δ3C-infected tumors expressed c-Myc and cyclin E at similar levels compared to WT-infected tumors, allowing cells to at least partially bypass p16-mediated cell cycle inhibition. The anti-apoptotic proteins, BCL2 and IRF4, were expressed in Δ3C-infected tumors, likely helping cells avoid c-Myc-induced apoptosis. Unexpectedly, Δ3C-infected tumors had increased T-cell infiltration, increased expression of T-cell chemokines (CCL5, CCL20 and CCL22) and enhanced type I interferon response in comparison to WT tumors. Together, these results reveal that EBNA3C contributes to, but is not essential for, EBV-induced lymphomagenesis in CBH mice, and suggest potentially important immunologic roles of EBNA3C in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Johannsen EC, Kaye KM. Epstein-Barr (infectious mononucleosis, Epstein-Barr virus-associated malignant diseases, and other diseases). Princ Pract Infect Dis. 2014;2: 1989–2010. 10.1016/B978-1-4557-4801-3.00141–7 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

