A zinc-finger fusion protein refines Gal4-defined neural circuits
- PMID: 30126464
- PMCID: PMC6102859
- DOI: 10.1186/s13041-018-0390-7
A zinc-finger fusion protein refines Gal4-defined neural circuits
Abstract
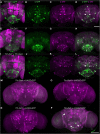
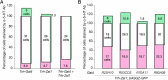
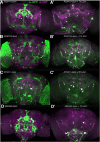
The analysis of behavior requires that the underlying neuronal circuits are identified and genetically isolated. In several major model species-most notably Drosophila-neurogeneticists identify and isolate neural circuits with a binary heterologous expression-control system: Gal4-UASG. One limitation of Gal4-UASG is that expression patterns are often too broad to map circuits precisely. To help refine the range of Gal4 lines, we developed an intersectional genetic AND operator. Interoperable with Gal4, the new system's key component is a fusion protein in which the DNA-binding domain of Gal4 has been replaced with a zinc finger domain with a different DNA-binding specificity. In combination with its cognate binding site (UASZ) the zinc-finger-replaced Gal4 ('Zal1') was functional as a standalone transcription factor. Zal1 transgenes also refined Gal4 expression ranges when combined with UASGZ, a hybrid upstream activation sequence. In this way, combining Gal4 and Zal1 drivers captured restricted cell sets compared with single drivers and improved genetic fidelity. This intersectional genetic AND operation presumably derives from the action of a heterodimeric transcription factor: Gal4-Zal1. Configurations of Zal1-UASZ and Zal1-Gal4-UASGZ are versatile tools for defining, refining, and manipulating targeted neural expression patterns with precision.
Conflict of interest statement
Ethics approval and consent to participate
No ethics approval was required or sought.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
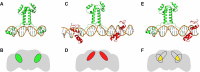
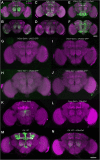
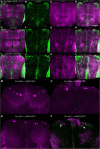
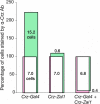
Similar articles
-
Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila.Nucleic Acids Res. 2016 Sep 6;44(15):7228-41. doi: 10.1093/nar/gkw371. Epub 2016 May 2. Nucleic Acids Res. 2016. PMID: 27137890 Free PMC article.
-
Facilitating Neuron-Specific Genetic Manipulations in Drosophila melanogaster Using a Split GAL4 Repressor.Genetics. 2017 Jun;206(2):775-784. doi: 10.1534/genetics.116.199687. Epub 2017 Mar 31. Genetics. 2017. PMID: 28363977 Free PMC article.
-
Focusing transgene expression in Drosophila by coupling Gal4 with a novel split-LexA expression system.Genetics. 2011 May;188(1):229-33. doi: 10.1534/genetics.110.126193. Epub 2011 Mar 2. Genetics. 2011. PMID: 21368278 Free PMC article.
-
The GAL4 System: A Versatile System for the Manipulation and Analysis of Gene Expression.Methods Mol Biol. 2016;1478:33-52. doi: 10.1007/978-1-4939-6371-3_2. Methods Mol Biol. 2016. PMID: 27730574 Review.
-
The Q-System: A Versatile Expression System for Drosophila.Methods Mol Biol. 2016;1478:53-78. doi: 10.1007/978-1-4939-6371-3_3. Methods Mol Biol. 2016. PMID: 27730575 Free PMC article. Review.
Cited by
-
Jatamansinol from Nardostachys jatamansi Ameliorates Tau-Induced Neurotoxicity in Drosophila Alzheimer's Disease Model.Mol Neurobiol. 2022 Oct;59(10):6091-6106. doi: 10.1007/s12035-022-02964-7. Epub 2022 Jul 22. Mol Neurobiol. 2022. PMID: 35864434
-
Anatomy and Neural Pathways Modulating Distinct Locomotor Behaviors in Drosophila Larva.Biology (Basel). 2021 Jan 25;10(2):90. doi: 10.3390/biology10020090. Biology (Basel). 2021. PMID: 33504061 Free PMC article. Review.
-
Serotonergic control of feeding microstructure in Drosophila.Front Behav Neurosci. 2023 Jan 17;16:1105579. doi: 10.3389/fnbeh.2022.1105579. eCollection 2022. Front Behav Neurosci. 2023. PMID: 36733453 Free PMC article.
-
2,5-Bis(2,2,2-trifluoroethoxy)phenyl-tethered 1,3,4-Oxadiazoles Derivatives: Synthesis, In Silico Studies, and Biological Assessment as Potential Candidates for Anti-Cancer and Anti-Diabetic Agent.Molecules. 2022 Dec 8;27(24):8694. doi: 10.3390/molecules27248694. Molecules. 2022. PMID: 36557829 Free PMC article.
-
Dextran sodium sulfate alters antioxidant status in the gut affecting the survival of Drosophila melanogaster.3 Biotech. 2022 Oct;12(10):280. doi: 10.1007/s13205-022-03349-2. Epub 2022 Sep 16. 3 Biotech. 2022. PMID: 36275361 Free PMC article.
References
-
- Altman D, et al. Statistics with confidence: confidence interval and statistical guidelines. Bristol: BMJ Books; 2000.
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

