Circadian clock protein BMAL1 regulates IL-1β in macrophages via NRF2
- PMID: 30127006
- PMCID: PMC6130388
- DOI: 10.1073/pnas.1800431115
Circadian clock protein BMAL1 regulates IL-1β in macrophages via NRF2
Abstract
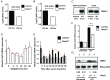
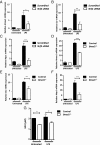
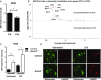
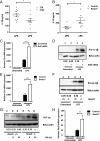
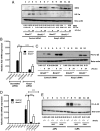
A variety of innate immune responses and functions are dependent on time of day, and many inflammatory conditions are associated with dysfunctional molecular clocks within immune cells. However, the functional importance of these innate immune clocks has yet to be fully characterized. NRF2 plays a critical role in the innate immune system, limiting inflammation via reactive oxygen species (ROS) suppression and direct repression of the proinflammatory cytokines, IL-1β and IL-6. Here we reveal that the core molecular clock protein, BMAL1, controls the mRNA expression of Nrf2 via direct E-box binding to its promoter to regulate its activity. Deletion of Bmal1 decreased the response of NRF2 to LPS challenge, resulting in a blunted antioxidant response and reduced synthesis of glutathione. ROS accumulation was increased in Bmal1-/- macrophages, facilitating accumulation of the hypoxic response protein, HIF-1α. Increased ROS and HIF-1α levels, as well as decreased activity of NRF2 in cells lacking BMAL1, resulted in increased production of the proinflammatory cytokine, IL-1β. The excessive prooxidant and proinflammatory phenotype of Bmal1-/- macrophages was rescued by genetic and pharmacological activation of NRF2, or through addition of antioxidants. Our findings uncover a clear role for the molecular clock in regulating NRF2 in innate immune cells to control the inflammatory response. These findings provide insights into the pathology of inflammatory conditions, in which the molecular clock, oxidative stress, and IL-1β are known to play a role.
Keywords: BMAL1; circadian clock; inflammation; macrophage; oxidative stress.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

