Structural determinants of 5-HT2B receptor activation and biased agonism
- PMID: 30127358
- PMCID: PMC6237183
- DOI: 10.1038/s41594-018-0116-7
Structural determinants of 5-HT2B receptor activation and biased agonism
Abstract
Serotonin (5-hydroxytryptamine; 5-HT) receptors modulate a variety of physiological processes ranging from perception, cognition and emotion to vascular and smooth muscle contraction, platelet aggregation, gastrointestinal function and reproduction. Drugs that interact with 5-HT receptors effectively treat diseases as diverse as migraine headaches, depression and obesity. Here we present four structures of a prototypical serotonin receptor-the human 5-HT2B receptor-in complex with chemically and pharmacologically diverse drugs, including methysergide, methylergonovine, lisuride and LY266097. A detailed analysis of these structures complemented by comprehensive interrogation of signaling illuminated key structural determinants essential for activation. Additional structure-guided mutagenesis experiments revealed binding pocket residues that were essential for agonist-mediated biased signaling and β-arrestin2 translocation. Given the importance of 5-HT receptors for a large number of therapeutic indications, insights derived from these studies should accelerate the design of safer and more effective medications.
Conflict of interest statement
Competing financial interests
The authors declare no competing financial interests
Figures
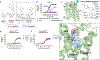
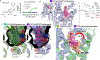
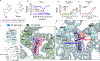


References
-
- Allen JA & Roth BL Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol 51, 117–44 (2011). - PubMed
-
- Vickers SP, Clifton PG, Dourish CT & Tecott LH Reduced satiating effect of d-fenfluramine in serotonin 5-HT(2C) receptor mutant mice. Psychopharmacology (Berl) 143, 309–14 (1999). - PubMed
-
- Roth BL Drugs and valvular heart disease. N Engl J Med 356, 6–9 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

