Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa
- PMID: 30127495
- PMCID: PMC6787116
- DOI: 10.1038/s41564-018-0217-4
Dynamics of antimicrobial resistance in intestinal Escherichia coli from children in community settings in South Asia and sub-Saharan Africa
Abstract
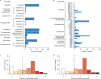
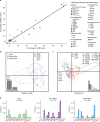
The dynamics of antimicrobial resistance (AMR) in developing countries are poorly understood, especially in community settings, due to a sparsity of data on AMR prevalence and genetics. We used a combination of phenotyping, genomics and antimicrobial usage data to investigate patterns of AMR amongst atypical enteropathogenic Escherichia coli (aEPEC) strains isolated from children younger than five years old in seven developing countries (four in sub-Saharan Africa and three in South Asia) over a three-year period. We detected high rates of AMR, with 65% of isolates displaying resistance to three or more drug classes. Whole-genome sequencing revealed a diversity of known genetic mechanisms for AMR that accounted for >95% of phenotypic resistance, with comparable rates amongst aEPEC strains associated with diarrhoea or asymptomatic carriage. Genetic determinants of AMR were associated with the geographic location of isolates, not E. coli lineage, and AMR genes were frequently co-located, potentially enabling the acquisition of multi-drug resistance in a single step. Comparison of AMR with antimicrobial usage data showed that the prevalence of resistance to fluoroquinolones and third-generation cephalosporins was correlated with usage, which was higher in South Asia than in Africa. This study provides much-needed insights into the frequency and mechanisms of AMR in intestinal E. coli in children living in community settings in developing countries.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. - PubMed
-
- Afset JE, Bevanger L, Romundstad P, Bergh K. Association of atypical enteropathogenic Escherichia coli (EPEC) with prolonged diarrhoea. J. Med. Microbiol. 2004;53:1137–1144. - PubMed
-
- Vieira MA, et al. Atypical enteropathogenic Escherichia coli as aetiologic agents of sporadic and outbreak-associated diarrhoea in Brazil. J. Med. Microbiol. 2016;65:998–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

