Screening and identification of key biomarkers in bladder carcinoma: Evidence from bioinformatics analysis
- PMID: 30127900
- PMCID: PMC6096082
- DOI: 10.3892/ol.2018.9002
Screening and identification of key biomarkers in bladder carcinoma: Evidence from bioinformatics analysis
Abstract
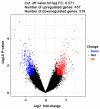
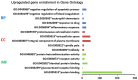
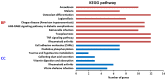
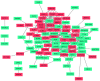
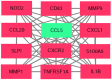
Bladder cancer (BC) is one of the most common urogenital malignancies. However, present studies of its multiple gene interaction and cellular pathways remain unable to accurately verify the genesis and the development of BC. The aim of the present study was to investigate the genetic signatures of BC and identify its potential molecular mechanisms. The gene expression profiles of GSE31189 were downloaded from the Gene Expression Omnibus database. The GSE31189 dataset contained 92 samples, including 52 BC and 40 non-cancerous urothelial cells. To further examine the biological functions of the identified differentially expressed genes (DEGs), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analyses were performed, and a protein-protein interaction (PPI) network was mapped using Cytoscape software. In total, 976 DEGs were identified in BC, including 457 upregulated genes and 519 downregulated genes. GO and KEGG pathway enrichment analyses indicated that upregulated genes were significantly enriched in the cell cycle and the negative regulation of the apoptotic process, while the downregulated genes were mainly involved in cell proliferation, cell adhesion molecules and oxidative phosphorylation pathways (P<0.05). From the PPI network, the 12 nodes with the highest degrees were screened as hub genes; these genes were involved in certain pathways, including the chemokine-mediated signaling pathway, fever generation, inflammatory response and the immune response nucleotide oligomerization domain-like receptor signaling pathway. The present study used bioinformatics analysis of gene profile datasets and identified potential therapeutic targets for BC.
Keywords: bladder cancer; differentially expressed genes; microarray analysis; protein-protein interaction network.
Figures



Similar articles
-
Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma.Mol Med Rep. 2018 Nov;18(5):4185-4196. doi: 10.3892/mmr.2018.9411. Epub 2018 Aug 21. Mol Med Rep. 2018. PMID: 30132538 Free PMC article.
-
The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data.Med Oncol. 2017 Jun;34(6):101. doi: 10.1007/s12032-017-0963-9. Epub 2017 Apr 21. Med Oncol. 2017. PMID: 28432618 Free PMC article.
-
Identification of key pathways and genes in nasopharyngeal carcinoma using bioinformatics analysis.Oncol Lett. 2019 May;17(5):4683-4694. doi: 10.3892/ol.2019.10133. Epub 2019 Mar 8. Oncol Lett. 2019. PMID: 30988824 Free PMC article.
-
Identification of differentially expressed genes and biological pathways in bladder cancer.Mol Med Rep. 2018 May;17(5):6425-6434. doi: 10.3892/mmr.2018.8711. Epub 2018 Mar 9. Mol Med Rep. 2018. PMID: 29532898 Free PMC article.
-
Identification of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis.Mol Med Rep. 2018 Aug;18(2):1538-1550. doi: 10.3892/mmr.2018.9095. Epub 2018 May 29. Mol Med Rep. 2018. PMID: 29845250 Free PMC article.
Cited by
-
Artificial intelligence-driven consensus gene signatures for improving bladder cancer clinical outcomes identified by multi-center integration analysis.Mol Oncol. 2022 Dec;16(22):4023-4042. doi: 10.1002/1878-0261.13313. Epub 2022 Sep 22. Mol Oncol. 2022. PMID: 36083778 Free PMC article.
-
Association of Mps one binder kinase activator 1 (MOB1) expression with poor disease-free survival in individuals with non-small cell lung cancer.Thorac Cancer. 2020 Oct;11(10):2830-2839. doi: 10.1111/1759-7714.13608. Epub 2020 Aug 25. Thorac Cancer. 2020. PMID: 32841529 Free PMC article.
-
Systematic analysis of the lysine malonylome in Sanghuangporus sanghuang.BMC Genomics. 2021 Nov 19;22(1):840. doi: 10.1186/s12864-021-08120-0. BMC Genomics. 2021. PMID: 34798813 Free PMC article.
-
Intersectin 1 (ITSN1) identified by comprehensive bioinformatic analysis and experimental validation as a key candidate biological target in breast cancer.Onco Targets Ther. 2019 Aug 30;12:7079-7093. doi: 10.2147/OTT.S216286. eCollection 2019. Onco Targets Ther. 2019. PMID: 31564893 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
