Determinants of Oligonucleotide Selectivity of APOBEC3B
- PMID: 30130104
- PMCID: PMC6644697
- DOI: 10.1021/acs.jcim.8b00427
Determinants of Oligonucleotide Selectivity of APOBEC3B
Abstract
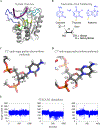
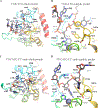
APOBEC3B (A3B) is a prominent source of mutation in many cancers. To date, it has been difficult to capture the native protein-DNA interactions that confer A3B's substrate specificity by crystallography due to the highly dynamic nature of wild-type A3B active site. We use computational tools to restore a recent crystal structure of a DNA-bound A3B C-terminal domain mutant construct to its wild type sequence, and run molecular dynamics simulations to study its substrate recognition mechanisms. Analysis of these simulations reveal dynamics of the native A3Bctd-oligonucleotide interactions, including the experimentally inaccessible loop 1-oligonucleotide interactions. A second series of simulations in which the target cytosine nucleotide was computationally mutated from a deoxyribose to a ribose show a change in sugar ring pucker, leading to a rearrangement of the binding site and revealing a potential intermediate in the binding pathway. Finally, apo simulations of A3B, starting from the DNA-bound open state, experience a rapid and consistent closure of the binding site, reaching conformations incompatible with substrate binding. This study reveals a more realistic and dynamic view of the wild type A3B binding site and provides novel insights for structure-guided design efforts for A3B.
Conflict of interest statement
Conflict of Interest Statement:
DAH & RSH are co-founders, consultants, and shareholders of ApoGen Biotechnologies, Inc. REA & HA are consultants of ApoGen Biotechnologies, Inc.
REA is a cofounder of, has equity interest in, and is on the scientific advisory board of Actavalon, Inc.
Figures

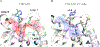



References
-
- Malim MH; Emerman M, HIV-1 Accessory Proteins--Ensuring Viral Survival in a Hostile Environment. Cell Host Microbe 2008, 3, 388–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

