Unraveling the structural and molecular properties of 34-residue levans with various branching degrees by replica exchange molecular dynamics simulations
- PMID: 30130368
- PMCID: PMC6103501
- DOI: 10.1371/journal.pone.0202578
Unraveling the structural and molecular properties of 34-residue levans with various branching degrees by replica exchange molecular dynamics simulations
Abstract
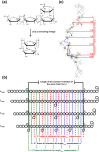
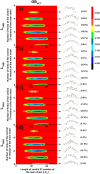
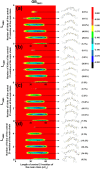
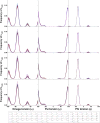
Levan has various potential applications in the pharmaceutical and food industries, such as cholesterol-lowering agents and prebiotics, due to its beneficial properties, which depend on its length and branching degree. A previous study also found that the branching degree of levan affected anti-tumor activities against SNU-1 and HepG2 tumor cell lines. Despite its promising potential, the properties of levans with different branching degrees are not well understood at the molecular level. In two models of the generalized Born implicit solvent (GBHCT and GBOBC1), we employed replica-exchange molecular dynamics simulations to explore conformational spaces of 34-residue levans (L34) with branching degrees of zero (LFO34B0), one (LFO34B1), three (LFO34B3) and five (LFO34B5), as well as to elucidate their structural and molecular properties. To ensure a fair comparison of the effects of branching degree on these properties, we focused on analyzing the properties of the central 21-residue of the main chains of all systems. Our results show that all major representative conformations tend to form helix-like structures with kinks, where two-kink helix-like structures have the highest population. As branching degree increases, the population of helix-like structures with zero or one kink tends to increase slightly. As the number of kinks in the structures with the same branching degree increases, the average values of the lengths and angles among centers of masses of three consecutive turns of residue i, i+3, and i+6 tended to decrease. Due to its highest occurring frequencies, the O6 (i)-H3O (i+1) hydrogen bond could be important for helix-like structure formation. Moreover, hydrogen bonds forming among the branching residue (br), branching position (bp) and other residues of L34B1, L34B3 and L34B5 were identified. The O1(bp)-H3O(br), O1(br)-H3O(br) and O5(br)-H1O(br) hydrogen bonds were found in the first-, second- and third-highest occurrence frequencies, respectively. Our study provides novel and important insights into conformational spaces and the structural and molecular properties of 34-residue levans with various branching degrees, which tend to form helix-like structures with kinks.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Replica exchange molecular dynamics simulations reveal the structural and molecular properties of levan-type fructo-oligosaccharides of various chain lengths.BMC Bioinformatics. 2016 Aug 17;17(1):306. doi: 10.1186/s12859-016-1182-7. BMC Bioinformatics. 2016. PMID: 27534934 Free PMC article.
-
Thermodynamics of helix formation in small peptides of varying length in vacuo, in implicit solvent, and in explicit solvent.J Mol Model. 2018 Dec 12;25(1):3. doi: 10.1007/s00894-018-3886-2. J Mol Model. 2018. PMID: 30542771
-
Effect of levan's branching structure on antitumor activity.Int J Biol Macromol. 2004 Jun;34(3):191-4. doi: 10.1016/j.ijbiomac.2004.04.001. Int J Biol Macromol. 2004. PMID: 15225991
-
[A turning point in the knowledge of the structure-function-activity relations of elastin].J Soc Biol. 2001;195(2):181-93. J Soc Biol. 2001. PMID: 11727705 Review. French.
-
Review of Levan polysaccharide: From a century of past experiences to future prospects.Biotechnol Adv. 2016 Sep-Oct;34(5):827-844. doi: 10.1016/j.biotechadv.2016.05.002. Epub 2016 May 10. Biotechnol Adv. 2016. PMID: 27178733 Review.
References
-
- Steinmetz M, Le Coq D, Aymerich S, Gonzy-Tréboul G, Gay P. The DNA sequence of the gene for the secreted Bacillus subtilis enzyme levansucrase and its genetic control sites. Mol Gen Genet. 1985; 200: 220–228. - PubMed
-
- Hettwer U, Jaeckel FR, Boch J, Meyer M, Rudolph K, Ullrich S. Cloning, Nucleotide Sequence, and Expression in Escherichia coli of Levansucrase Genes from the Plant Pathogens Pseudomonas syringae pv. glycinea and P. syringaepv. phaseolicola. Appl Environ Microbiol. 1998; 64: 3180–3187. - PMC - PubMed
-
- Song KB, Seo JW, Kim MG, Rhee SK. Levansucrase of Rahnella aquatilis ATCC33071: gene cloning, expression, and levan formation. Ann N Y Acad Sci. 1998; 864: 506–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

