Systematic Analysis of Survival-Associated Alternative Splicing Signatures in Gastrointestinal Pan-Adenocarcinomas
- PMID: 30131306
- PMCID: PMC6116578
- DOI: 10.1016/j.ebiom.2018.07.040
Systematic Analysis of Survival-Associated Alternative Splicing Signatures in Gastrointestinal Pan-Adenocarcinomas
Abstract
Background: Gastrointestinal pan-adenocarcinomas, which mainly include adenocarcinomas of the esophagus, stomach, colon, and rectum, place a heavy burden on society owing to their poor prognoses. Since aberrant alternative splicing (AS) are starting to be considered as efficacious signatures for tumor prognosis predicting and therapeutic targets, systematic analysis of AS events is urgent.
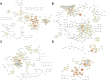
Methods: Prognosis-related AS events were selected by using univariate COX regression analysis. Gene functional enrichment analysis revealed the pathways enriched by prognosis-related AS. Then, prognostic signatures based on AS events were developed for prognosis prediction. Potential mechanism to regulate splicing events by splicing factors was analyzed via Pearson correlation and regulatory networks were constructed.
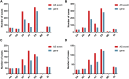
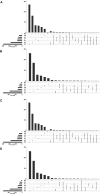
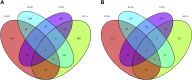
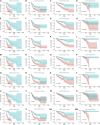
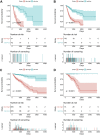
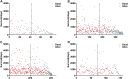
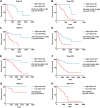
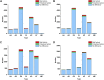
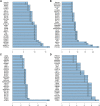
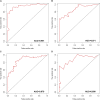
Findings: A total of 967, 918, 674, and 406 AS events were identified as prognosis-related AS events in esophagus, stomach, colon, and rectum adenocarcinomas, respectively. Survival-associated AS events were distinguishing in the four subtypes of adenocarcinoma. Furthermore, computational algorithm results indicated that perturbation of ribosome and ubiquitin-mediated proteolysis pathways were the potential molecular mechanisms corresponding to inferior prognoses. Most notably, several prognostic signatures based on AS events displayed moderate performance in prognosis predicting. The area under curve values of the time-dependent receiver operating characteristic were 0.961, 0.871, 0.870, and 0.890 in esophagus, stomach, colon, and rectum adenocarcinomas. Survival-associated splicing factors were submitted to construct the AS regulatory network, which could be an underlying mechanism of AS events.
Interpretation: AS may could be ideal indiactors in the prognosis of gastrointestinal pan-adenocarcinomas. Exploring interesting splicing regulatory networks is conducive to solve the puzzles of AS.
Keywords: Alternative splicing; Gastrointestinal pan-adenocarcinomas; Prognosis; Splicing factors.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Systemic characterization of alternative splicing related to prognosis and immune infiltration in malignant mesothelioma.BMC Cancer. 2021 Jul 22;21(1):848. doi: 10.1186/s12885-021-08548-3. BMC Cancer. 2021. PMID: 34294080 Free PMC article.
-
Survival-Associated Alternative Messenger RNA Splicing Signatures in Pancreatic Ductal Adenocarcinoma: A Study Based on RNA-Sequencing Data.DNA Cell Biol. 2019 Nov;38(11):1207-1222. doi: 10.1089/dna.2019.4862. Epub 2019 Sep 4. DNA Cell Biol. 2019. PMID: 31483163
-
Prognostic Signature of Alternative Splicing Events in Bladder Urothelial Carcinoma Based on Spliceseq Data from 317 Cases.Cell Physiol Biochem. 2018;48(3):1355-1368. doi: 10.1159/000492094. Epub 2018 Jul 26. Cell Physiol Biochem. 2018. PMID: 30048970
-
Clinical significance of molecular subtypes of gastrointestinal tract adenocarcinoma.World J Gastrointest Oncol. 2022 Mar 15;14(3):628-645. doi: 10.4251/wjgo.v14.i3.628. World J Gastrointest Oncol. 2022. PMID: 35321271 Free PMC article. Review.
-
Cancer Stem Cells in Tumor Microenvironment of Adenocarcinoma of the Stomach, Colon, and Rectum.Cancers (Basel). 2022 Aug 16;14(16):3948. doi: 10.3390/cancers14163948. Cancers (Basel). 2022. PMID: 36010940 Free PMC article. Review.
Cited by
-
Alternative splicing perturbation landscape identifies RNA binding proteins as potential therapeutic targets in cancer.Mol Ther Nucleic Acids. 2021 Apr 9;24:792-806. doi: 10.1016/j.omtn.2021.04.005. eCollection 2021 Jun 4. Mol Ther Nucleic Acids. 2021. PMID: 33996260 Free PMC article.
-
Characterization of alternative splicing events and prognostic signatures in breast cancer.BMC Cancer. 2021 May 22;21(1):587. doi: 10.1186/s12885-021-08305-6. BMC Cancer. 2021. PMID: 34022836 Free PMC article.
-
Comprehensive Analysis of Alternative Splicing Signature in Gastric Cancer Prognosis Based on The Cancer Genome Atlas (TCGA) and SpliceSeq Databases.Med Sci Monit. 2020 Nov 21;26:e925772. doi: 10.12659/MSM.925772. Med Sci Monit. 2020. PMID: 33219199 Free PMC article.
-
Novel Insights Into Triple-Negative Breast Cancer Prognosis by Comprehensive Characterization of Aberrant Alternative Splicing.Front Genet. 2020 Jun 11;11:534. doi: 10.3389/fgene.2020.00534. eCollection 2020. Front Genet. 2020. PMID: 32595697 Free PMC article.
-
Genome-wide analysis reveals the association between alternative splicing and DNA methylation across human solid tumors.BMC Med Genomics. 2020 Jan 6;13(1):4. doi: 10.1186/s12920-019-0654-9. BMC Med Genomics. 2020. PMID: 31906954 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

