High-Throughput Screening Identifies Genes Required for Candida albicans Induction of Macrophage Pyroptosis
- PMID: 30131363
- PMCID: PMC6106084
- DOI: 10.1128/mBio.01581-18
High-Throughput Screening Identifies Genes Required for Candida albicans Induction of Macrophage Pyroptosis
Abstract
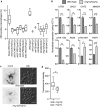
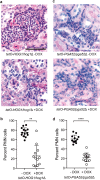
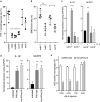
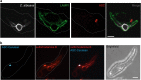
The innate immune system is the first line of defense against invasive fungal infections. As a consequence, many successful fungal pathogens have evolved elegant strategies to interact with host immune cells. For example, Candida albicans undergoes a morphogenetic switch coupled to cell wall remodeling upon phagocytosis by macrophages and then induces macrophage pyroptosis, an inflammatory cell death program. To elucidate the genetic circuitry through which C. albicans orchestrates this host response, we performed the first large-scale analysis of C. albicans interactions with mammalian immune cells. We identified 98 C. albicans genes that enable macrophage pyroptosis without influencing fungal cell morphology in the macrophage, including specific determinants of cell wall biogenesis and the Hog1 signaling cascade. Using these mutated genes, we discovered that defects in the activation of pyroptosis affect immune cell recruitment during infection. Examining host circuitry required for pyroptosis in response to C. albicans infection, we discovered that inflammasome priming and activation can be decoupled. Finally, we observed that
Keywords: Candida; cell wall remodelling; functional genomics; fungal morphogenesis; fungal pathogenesis; host-pathogen interaction; pyroptosis.
Copyright © 2018 O’Meara et al.
Figures



Comment in
-
Insights into the host-pathogen interaction: C. albicans manipulation of macrophage pyroptosis.Microb Cell. 2018 Nov 12;5(12):566-568. doi: 10.15698/mic2018.12.662. Microb Cell. 2018. PMID: 30533421 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
