Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells
- PMID: 30131803
- PMCID: PMC6090154
- DOI: 10.3389/fimmu.2018.01770
Armed and Ready: Transcriptional Regulation of Tissue-Resident Memory CD8 T Cells
Abstract
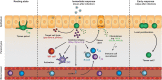
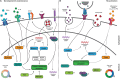
A fundamental benefit of immunological memory is the ability to respond in an enhanced manner upon secondary encounter with the same pathogen. Tissue-resident memory CD8 T (TRM) cells contribute to improved protection against reinfection through the generation of immediate effector responses at the site of pathogen entry. Key to the potential of TRM cells to develop rapid recall responses is their location within the epithelia of the skin, lungs, and intestines at prime entry sites of pathogens. TRM cells are among the first immune cells to respond to pathogens that have been previously encountered in an antigen-specific manner. Upon recognition of invading pathogens, TRM cells release IFN-γ and other pro-inflammatory cytokines and chemokines. These effector molecules activate the surrounding epithelial tissue and recruit other immune cells including natural killer (NK) cells, B cells, and circulating memory CD8 T cells to the site of infection. The repertoire of TRM effector functions also includes the direct lysis of infected cells through the release of cytotoxic molecules such as perforin and granzymes. The mechanisms enabling TRM cells to respond in such a rapid manner are gradually being uncovered. In this review, we will address the signals that instruct TRM generation and maintenance as well as the underlying transcriptional network that keeps TRM cells in a deployment-ready modus. Furthermore, we will discuss how TRM cells respond to reinfection of the tissue and how transcription factors may control immediate and proliferative TRM responses.
Keywords: BLIMP-1; Notch; RUNX3; T cell diferentiation; homolog of Blimp-1 in T cells; secondary responses; tissue-resident memory T cells; transcription factors.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

