Spindle Dynamics Model Explains Chromosome Loss Rates in Yeast Polyploid Cells
- PMID: 30131823
- PMCID: PMC6091489
- DOI: 10.3389/fgene.2018.00296
Spindle Dynamics Model Explains Chromosome Loss Rates in Yeast Polyploid Cells
Abstract
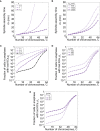
Faithful chromosome segregation, driven by the mitotic spindle, is essential for organismal survival. Neopolyploid cells from diverse species exhibit a significant increase in mitotic errors relative to their diploid progenitors, resulting in chromosome nondisjunction. In the model system Saccharomyces cerevisiae, the rate of chromosome loss in haploid and diploid cells is measured to be one thousand times lower than the rate of loss in isogenic tetraploid cells. Currently it is unknown what constrains the number of chromosomes that can be segregated with high fidelity in an organism. Here we developed a simple mathematical model to study how different rates of chromosome loss in cells with different ploidy can arise from changes in (1) spindle dynamics and (2) a maximum duration of mitotic arrest, after which cells enter anaphase. We apply this model to S. cerevisiae to show that this model can explain the observed rates of chromosome loss in S. cerevisiae cells of different ploidy. Our model describes how small increases in spindle assembly time can result in dramatic differences in the rate of chromosomes loss between cells of increasing ploidy and predicts the maximum duration of mitotic arrest.
Keywords: cell cycle regulation; chromosome loss; chromosome segregation; genome instability; polyploidy; spindle assembly; theoretical modeling.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

