An integrated clinical program and crowdsourcing strategy for genomic sequencing and Mendelian disease gene discovery
- PMID: 30131872
- PMCID: PMC6089983
- DOI: 10.1038/s41525-018-0060-9
An integrated clinical program and crowdsourcing strategy for genomic sequencing and Mendelian disease gene discovery
Abstract
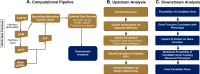
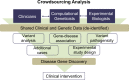
Despite major progress in defining the genetic basis of Mendelian disorders, the molecular etiology of many cases remains unknown. Patients with these undiagnosed disorders often have complex presentations and require treatment by multiple health care specialists. Here, we describe an integrated clinical diagnostic and research program using whole-exome and whole-genome sequencing (WES/WGS) for Mendelian disease gene discovery. This program employs specific case ascertainment parameters, a WES/WGS computational analysis pipeline that is optimized for Mendelian disease gene discovery with variant callers tuned to specific inheritance modes, an interdisciplinary crowdsourcing strategy for genomic sequence analysis, matchmaking for additional cases, and integration of the findings regarding gene causality with the clinical management plan. The interdisciplinary gene discovery team includes clinical, computational, and experimental biomedical specialists who interact to identify the genetic etiology of the disease, and when so warranted, to devise improved or novel treatments for affected patients. This program effectively integrates the clinical and research missions of an academic medical center and affords both diagnostic and therapeutic options for patients suffering from genetic disease. It may therefore be germane to other academic medical institutions engaged in implementing genomic medicine programs.
Conflict of interest statement
R.C.G. receives compensation for speaking or advisory services from AIA, Genome Medical, Helix, Invitae, Illumina, Prudential, and Roche. R.C.G., N.F., and S.R.S. are on the board of Veritas Genetics. C.A.M. receives compensation for advisory services from Personome and Recombine, and royalties for genetic testing from Partners HealthCare. The other authors declare no competing interests.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

