Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors
- PMID: 30133130
- PMCID: PMC6210052
- DOI: 10.1002/1878-0261.12375
Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors
Abstract
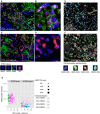
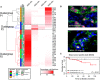
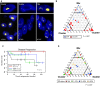
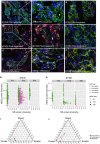
Targeted therapy for patients with HER2-positive (HER2+) breast cancer has improved overall survival, but many patients still suffer relapse and death from the disease. Intratumor heterogeneity of both estrogen receptor (ER) and HER2 expression has been proposed to play a key role in treatment failure, but little work has been done to comprehensively study this heterogeneity at the single-cell level. In this study, we explored the clinical impact of intratumor heterogeneity of ER protein expression, HER2 protein expression, and HER2 gene copy number alterations. Using combined immunofluorescence and in situ hybridization on tissue sections followed by a validated computational approach, we analyzed more than 13 000 single tumor cells across 37 HER2+ breast tumors. The samples were taken both before and after neoadjuvant chemotherapy plus HER2-targeted treatment, enabling us to study tumor evolution as well. We found that intratumor heterogeneity for HER2 copy number varied substantially between patient samples. Highly heterogeneous tumors were associated with significantly shorter disease-free survival and fewer long-term survivors. Patients for which HER2 characteristics did not change during treatment had a significantly worse outcome. This work shows the impact of intratumor heterogeneity in molecular diagnostics for treatment selection in HER2+ breast cancer patients and the power of computational scoring methods to evaluate in situ molecular markers in tissue biopsies.
Keywords: HER2; breast cancer; heterogeneity; in situ analysis; outcome; therapy response.
© 2018 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures



References
-
- Arena V, Pennacchia I, Vecchio FM and Carbone A (2013) HER‐2 intratumoral heterogeneity. Mod Pathol 26, 607–609. - PubMed
-
- Arnould L, Arveux P, Couturier J, Gelly‐Marty M, Loustalot C, Ettore F, Sagan C, Antoine M, Penault‐Llorca F, Vasseur B et al (2007) Pathologic complete response to trastuzumab‐based neoadjuvant therapy is related to the level of HER‐2 amplification. Clin Cancer Res 13, 6404–6409. - PubMed
-
- Ballard M, Jalikis F, Krings G, Schmidt RA, Chen YY, Rendi MH, Dintzis SM, Jensen KC, West RB, Sibley RK et al (2017) ‘Non‐classical’ HER2 FISH results in breast cancer: a multi‐institutional study. Mod Pathol 30, 227–235. - PubMed
-
- Bartlett AI, Starcyznski J, Robson T, Maclellan A, Campbell FM, van de Velde CJ, Hasenburg A, Markopoulos C, Seynaeve C, Rea D et al (2011) Heterogeneous HER2 gene amplification: impact on patient outcome and a clinically relevant definition. Am J Clin Pathol 136, 266–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

