Brr6 plays a role in gene recruitment and transcriptional regulation at the nuclear envelope
- PMID: 30133335
- PMCID: PMC6254580
- DOI: 10.1091/mbc.E18-04-0258
Brr6 plays a role in gene recruitment and transcriptional regulation at the nuclear envelope
Abstract
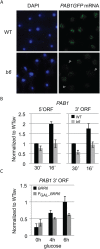
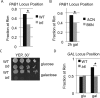
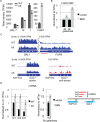
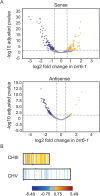
Correlation between transcriptional regulation and positioning of genes at the nuclear envelope is well established in eukaryotes, but the mechanisms involved are not well understood. We show that brr6-1, a mutant of the essential yeast envelope transmembrane protein Brr6p, impairs normal positioning and expression of the PAB1 and FUR4- GAL1,10,7 loci. Similarly, expression of a dominant negative nucleoplasmic Brr6 fragment in wild-type cells reproduced many of the brr6-1 effects. Histone chromatin immunoprecipitation (ChIP) experiments showed decreased acetylation at the key histone H4K16 residue in the FUR4-GAL1,10,7 region in brr6-1. Importantly, blocking deacetylation significantly suppressed selected brr6-1 phenotypes. ChIPseq with FLAG-tagged Brr6 fragments showed enrichment at FUR4 and several other genes that showed striking changes in brr6-1 RNAseq data. These associations depended on a Brr6 putative zinc finger domain. Importantly, artificially tethering the GAL1 locus to the envelope suppressed the brr6-1 effects on GAL1 and FUR4 expression and increased H4K16 acetylation between GAL1 and FUR4 in the mutant. Together these results argue that Brr6 interacts with chromatin, helping to maintain normal chromatin architecture and transcriptional regulation of certain loci at the nuclear envelope.
Figures


References
-
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. (1998). Perinuclear localization of chromatin facilitates transcriptional silencing. Nature , 592–595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

