Investigating genetic-and-epigenetic networks, and the cellular mechanisms occurring in Epstein-Barr virus-infected human B lymphocytes via big data mining and genome-wide two-sided NGS data identification
- PMID: 30133498
- PMCID: PMC6105016
- DOI: 10.1371/journal.pone.0202537
Investigating genetic-and-epigenetic networks, and the cellular mechanisms occurring in Epstein-Barr virus-infected human B lymphocytes via big data mining and genome-wide two-sided NGS data identification
Abstract
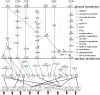
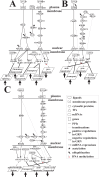
Epstein-Barr virus (EBV), also known as human herpesvirus 4, is prevalent in all human populations. EBV mainly infects human B lymphocytes and epithelial cells, and is therefore associated with their various malignancies. To unravel the cellular mechanisms during the infection, we constructed interspecies networks to investigate the molecular cross-talk mechanisms between human B cells and EBV at the first (0-24 hours) and second (8-72 hours) stages of EBV infection. We first constructed a candidate genome-wide interspecies genetic-and-epigenetic network (the candidate GIGEN) by big database mining. We then pruned false positives in the candidate GIGEN to obtain the real GIGENs at the first and second infection stages in the lytic phase by their corresponding next-generation sequencing data through dynamic interaction models, the system identification approach, and the system order detection method. The real GIGENs are very complex and comprise protein-protein interaction networks, gene/microRNA (miRNA)/long non-coding RNA regulation networks, and host-virus cross-talk networks. To understand the molecular cross-talk mechanisms underlying EBV infection, we extracted the core GIGENs including host-virus core networks and host-virus core pathways from the real GIGENs using the principal network projection method. According to the results, we found that the activities of epigenetics-associated human proteins or genes were initially inhibited by viral proteins and miRNAs, and human immune responses were then dysregulated by epigenetic modification. We suggested that EBV exploits viral proteins and miRNAs, such as EBNA1, BPLF1, BALF3, BVRF1 and miR-BART14, to develop its defensive mechanism to defeat multiple immune attacks by the human immune system, promotes virion production, and facilitates the transportation of viral particles by activating the human genes NRP1 and CLIC5. Ultimately, we propose a therapeutic intervention comprising thymoquinone, valpromide, and zebularine to act as inhibitors of EBV-associated malignancies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes.J Virol. 2019 Jun 14;93(13):e00226-19. doi: 10.1128/JVI.00226-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019051 Free PMC article.
-
Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus.mBio. 2021 Mar 30;12(2):e03440-20. doi: 10.1128/mBio.03440-20. mBio. 2021. PMID: 33785626 Free PMC article.
-
MicroRNAs of Epstein-Barr Virus Attenuate T-Cell-Mediated Immune Control In Vivo.mBio. 2019 Jan 15;10(1):e01941-18. doi: 10.1128/mBio.01941-18. mBio. 2019. PMID: 30647153 Free PMC article.
-
Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases.Front Immunol. 2020 Mar 3;11:367. doi: 10.3389/fimmu.2020.00367. eCollection 2020. Front Immunol. 2020. PMID: 32194570 Free PMC article. Review.
-
Regulation and dysregulation of Epstein-Barr virus latency: implications for the development of autoimmune diseases.Autoimmunity. 2008 May;41(4):298-328. doi: 10.1080/08916930802024772. Autoimmunity. 2008. PMID: 18432410 Review.
Cited by
-
Systems Approach to Pathogenic Mechanism of Type 2 Diabetes and Drug Discovery Design Based on Deep Learning and Drug Design Specifications.Int J Mol Sci. 2020 Dec 26;22(1):166. doi: 10.3390/ijms22010166. Int J Mol Sci. 2020. PMID: 33375269 Free PMC article.
-
Identifying Drug Targets of Oral Squamous Cell Carcinoma through a Systems Biology Method and Genome-Wide Microarray Data for Drug Discovery by Deep Learning and Drug Design Specifications.Int J Mol Sci. 2022 Sep 8;23(18):10409. doi: 10.3390/ijms231810409. Int J Mol Sci. 2022. PMID: 36142321 Free PMC article.
-
Network Approaches for Charting the Transcriptomic and Epigenetic Landscape of the Developmental Origins of Health and Disease.Genes (Basel). 2022 Apr 26;13(5):764. doi: 10.3390/genes13050764. Genes (Basel). 2022. PMID: 35627149 Free PMC article. Review.
-
Long Non-coding RNAs in Gammaherpesvirus Infections: Their Roles in Tumorigenic Mechanisms.Front Microbiol. 2021 Jan 15;11:604536. doi: 10.3389/fmicb.2020.604536. eCollection 2020. Front Microbiol. 2021. PMID: 33519750 Free PMC article. Review.
-
Landscape of the Epstein-Barr virus-host chromatin interactome and gene regulation.EMBO J. 2025 Jul;44(13):3872-3915. doi: 10.1038/s44318-025-00466-5. Epub 2025 May 27. EMBO J. 2025. PMID: 40425856 Free PMC article.
References
-
- Epstein MA, Barr YM. Cultivation in Vitro of Human Lymphoblasts from Burkitts Malignant Lymphoma. Lancet. 1964;1(732):252-&. PubMed PMID: WOS:A19647819B00199. - PubMed
-
- Cen H, McKnight JL. EBV-immortalized isogenic human B-cell clones exhibit differences in DNA-protein complex formation on the BZLF1 and BRLF1 promoter regions among latent, lytic and TPA-activated cell lines. Virus Res. 1994;31(1):89–107. Epub 1994/01/01. . - PubMed
-
- Hong GK, Delecluse H- J, Gruffat H, Morrison TE, Feng W- H, Sergeant A, et al. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. Journal of virology. 2004;78(10):4983–92. 10.1128/JVI.78.10.4983-4992.2004 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

