Acceleration of biomolecular kinetics in Gaussian accelerated molecular dynamics
- PMID: 30134710
- PMCID: PMC6901173
- DOI: 10.1063/1.5024217
Acceleration of biomolecular kinetics in Gaussian accelerated molecular dynamics
Abstract
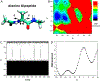
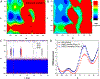
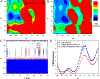
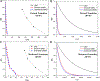
Recent studies demonstrated that Gaussian accelerated molecular dynamics (GaMD) is a robust computational technique, which provides simultaneous unconstrained enhanced sampling and free energy calculations of biomolecules. However, the exact acceleration of biomolecular dynamics or speedup of kinetic rates in GaMD simulations and, more broadly, in enhanced sampling methods, remains a challenging task to be determined. Here, the GaMD acceleration is examined using alanine dipeptide in explicit solvent as a biomolecular model system. Relative to long conventional molecular dynamics simulation, GaMD simulations exhibited ∼36-67 times speedup for sampling of the backbone dihedral transitions. The acceleration depended on level of the GaMD boost potential. Furthermore, Kramers' rate theory was applied to estimate GaMD acceleration using simulation-derived diffusion coefficients, curvatures and barriers of free energy profiles. In most cases, the calculations also showed significant speedup of dihedral transitions in GaMD, although the GaMD acceleration factors tended to be underestimated by ∼3-96 fold. Because greater boost potential can be applied in GaMD simulations of systems with increased sizes, which potentially leads to higher acceleration, it is subject to future studies on accelerating the dynamics and recovering kinetic rates of larger biomolecules such as proteins and protein-protein/nucleic acid complexes.
Figures
Similar articles
-
Multiple Parameter Replica Exchange Gaussian Accelerated Molecular Dynamics for Enhanced Sampling and Free Energy Calculation of Biomolecular Systems.J Chem Theory Comput. 2024 Aug 13;20(15):6485-6499. doi: 10.1021/acs.jctc.4c00501. Epub 2024 Jul 31. J Chem Theory Comput. 2024. PMID: 39085770
-
Gaussian Accelerated Molecular Dynamics in NAMD.J Chem Theory Comput. 2017 Jan 10;13(1):9-19. doi: 10.1021/acs.jctc.6b00931. Epub 2016 Dec 30. J Chem Theory Comput. 2017. PMID: 28034310 Free PMC article.
-
Replica Exchange Gaussian Accelerated Molecular Dynamics: Improved Enhanced Sampling and Free Energy Calculation.J Chem Theory Comput. 2018 Apr 10;14(4):1853-1864. doi: 10.1021/acs.jctc.7b01226. Epub 2018 Mar 12. J Chem Theory Comput. 2018. PMID: 29489349 Free PMC article.
-
Gaussian accelerated molecular dynamics for elucidation of drug pathways.Expert Opin Drug Discov. 2018 Nov;13(11):1055-1065. doi: 10.1080/17460441.2018.1538207. Epub 2018 Oct 29. Expert Opin Drug Discov. 2018. PMID: 30371112 Free PMC article. Review.
-
Binding Analysis Using Accelerated Molecular Dynamics Simulations and Future Perspectives.Adv Appl Bioinform Chem. 2022 Jan 6;15:1-19. doi: 10.2147/AABC.S247950. eCollection 2022. Adv Appl Bioinform Chem. 2022. PMID: 35023931 Free PMC article. Review.
Cited by
-
Most Probable Druggable Pockets in Mutant p53-Arg175His Clusters Extracted from Gaussian Accelerated Molecular Dynamics Simulations.Protein J. 2022 Feb;41(1):27-43. doi: 10.1007/s10930-022-10041-0. Epub 2022 Jan 31. Protein J. 2022. PMID: 35099676
-
Enhanced-Sampling Simulations for the Estimation of Ligand Binding Kinetics: Current Status and Perspective.Front Mol Biosci. 2022 Jun 8;9:899805. doi: 10.3389/fmolb.2022.899805. eCollection 2022. Front Mol Biosci. 2022. PMID: 35755817 Free PMC article. Review.
-
G-Protein-Coupled Receptor-Membrane Interactions Depend on the Receptor Activation State.J Comput Chem. 2020 Feb 15;41(5):460-471. doi: 10.1002/jcc.26082. Epub 2019 Oct 10. J Comput Chem. 2020. PMID: 31602675 Free PMC article.
-
15 Years of molecular simulation of drug-binding kinetics.Expert Opin Drug Discov. 2023 Jul-Dec;18(12):1333-1348. doi: 10.1080/17460441.2023.2264770. Epub 2023 Nov 1. Expert Opin Drug Discov. 2023. PMID: 37789731 Free PMC article. Review.
-
Mechanism of Ligand Binding to Theophylline RNA Aptamer.J Chem Inf Model. 2024 Feb 12;64(3):1017-1029. doi: 10.1021/acs.jcim.3c01454. Epub 2024 Jan 16. J Chem Inf Model. 2024. PMID: 38226603 Free PMC article.
References
-
- Schuetz DA, de Witte WEA, Wong YC, Knasmueller B, Richter L, Kokh DB, Sadiq SK, Bosma R, Nederpelt I, Heitman LH, Segala E, Amaral M, Guo D, Andres D, Georgi V, Stoddart LA, Hill S, Cooke RM, De Graaf C, Leurs R, Frech M, Wade RC, de Lange ECM, IJzerman AP, Muller-Fahrnow A, and Ecker GF, Drug Discov Today 22 (2017) 896. - PubMed
-
- Henzler-Wildman K, and Kern D, Nature 450 (2007) 964. - PubMed
-
- Vural D, Hu X, Lindner B, Jain N, Miao Y, Cheng X, Liu Z, Hong L, and Smith JC, Biochimica et biophysica acta 1861 (2017) 3638. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

