The microbiota in the intestinal and respiratory tracts of naked mole-rats revealed by high-throughput sequencing
- PMID: 30134830
- PMCID: PMC6103993
- DOI: 10.1186/s12866-018-1226-4
The microbiota in the intestinal and respiratory tracts of naked mole-rats revealed by high-throughput sequencing
Abstract
Background: The naked mole-rat (NMR, Heterocephalus glaber) is being bred as a novel laboratory animal due to its unique biological characteristics, including longevity, cancer resistance, hypoxia tolerance, and pain insensitivity. It is expected that differences exist between the microbiota of wild NMRs and that of NMRs in an artificial environment. Overall, the effect of environment on changes in the NMR microbiota remains unknown. In an attempt to understand the microbiota composition of NMRs in captivity, variability in the microbiota of the intestinal and respiratory tracts of two groups of NMRs was assessed under two conditions.
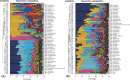
Results: The results obtained by high-throughput sequencing revealed significant differences at the phylum, class, order, family and genus levels in the microbiota between the two groups of NMRs examined (first group in conventional environment, second group in barrier environment). For the trachea, 24 phyla and 533 genera and 26 phyla and 733 genera were identified for the first and second groups of animals. Regarding the cecum, 23 phyla and 385 genera and 25 phyla and 110 genera were identified in the microbiota of first and second groups of animals. There were no obvious differences between females and males or young and adult animals.
Conclusions: Our results suggest that the intestinal and respiratory tract NMR microbiota changed during captivity, which may be related to the transition to the breeding environment. Such changes in the microbiota of NMRs may have an effect on the original characteristics, which may be the direction of further research studies.
Keywords: Bacterial diversity; Cecum; Microbiota; Naked mole-rats; Trachea.
Conflict of interest statement
Ethics approval and consent to participate
The animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the Second Military Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

