Deciphering the oviductal extracellular vesicles content across the estrous cycle: implications for the gametes-oviduct interactions and the environment of the potential embryo
- PMID: 30134841
- PMCID: PMC6103977
- DOI: 10.1186/s12864-018-4982-5
Deciphering the oviductal extracellular vesicles content across the estrous cycle: implications for the gametes-oviduct interactions and the environment of the potential embryo
Abstract
Background: The success of early reproductive events depends on an appropriate communication between gametes/embryos and the oviduct. Extracellular vesicles (EVs) contained in oviductal secretions have been suggested as new players in mediating this crucial cross-talk by transferring their cargo (proteins, mRNA and small ncRNA) from cell to cell. However, little is known about the oviductal EVs (oEVS) composition and their implications in the reproductive success. The aim of the study was to determine the oEVs content at protein, mRNA and small RNA level and to examine whether the oEVs content is under the hormonal influence of the estrous cycle.
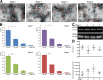
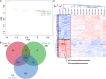
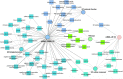
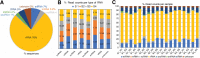
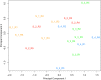
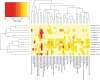
Results: We identified the presence of oEVs, exosomes and microvesicles, in the bovine oviductal fluid at different stages of the estrous cycle (postovulatory-stage, early luteal phase, late luteal phase and pre-ovulatory stage) and demonstrated that their composition is under hormonal regulation. RNA-sequencing identified 903 differentially expressed transcripts (FDR < 0.001) in oEVs across the estrous cycle. Moreover, small RNA-Seq identified the presence of different types of ncRNAs (miRNAs, rRNA fragments, tRNA fragments, snRNA, snoRNA, and other ncRNAs), which were partially also under hormonal influence. Major differences were found between post-ovulatory and the rest of the stages analyzed for mRNAs. Interesting miRNAs identified in oEVs and showing differential abundance among stages, miR-34c and miR-449a, have been associated with defective cilia in the oviduct and infertility. Furthermore, functional annotation of the differentially abundant mRNAs identified functions related to exosome/vesicles, cilia expression, embryo development and many transcripts encoding ribosomal proteins. Moreover, the analysis of oEVs protein content also revealed changes across the estrous cycle. Mass spectrometry identified 336 clusters of proteins in oEVs, of which 170 were differentially abundant across the estrous cycle (p-value< 0.05, ratio < 0.5 or ratio > 2). Our data revealed proteins related to early embryo development and gamete-oviduct interactions as well as numerous ribosomal proteins.
Conclusions: Our study provides with the first molecular signature of oEVs across the bovine estrous cycle, revealing marked differences between post- and pre-ovulatory stages. Our findings contribute to a better understanding of the potential role of oEVs as modulators of gamete/embryo-maternal interactions and their implications for the reproductive success.
Keywords: Exosomes; Gamete/embryo-maternal interaction; Hormones; Microvesicles; Oviduct; Proteomics; RNA-sequencing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- da Silveira JC, Veeramachaneni DN, Winger QA, Carnevale EM, Bouma GJ. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: a possible new form of cell communication within the ovarian follicle. Biol Reprod. 2012;86(3):71. doi: 10.1095/biolreprod.111.093252. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- n° 267196./EU Marie-Curie FP7 COFUND People Programme, AgreenSkills fellowship
- no.3569/SMHARTproject, by the European Regional Development Fund (ERDF), the Conseil Régional du Centre, the French National Institute for Agricultural Research (INRA) and the French National Institute of Health and Medical Research (Inserm).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

