Pathogenic variability among Pasteurella multocida type A isolates from Brazilian pig farms
- PMID: 30134904
- PMCID: PMC6103967
- DOI: 10.1186/s12917-018-1565-2
Pathogenic variability among Pasteurella multocida type A isolates from Brazilian pig farms
Abstract
Background: Pasteurella multocida type A (PmA) is considered a secondary agent of pneumonia in pigs. The role of PmA as a primary pathogen was investigated by challenging pigs with eight field strains isolated from pneumonia and serositis in six Brazilian states. Eight groups of eight pigs each were intranasally inoculated with different strains of PmA (1.5 mL/nostril of 10e7 CFU/mL). The control group (n = 12) received sterile PBS. The pigs were euthanized by electrocution and necropsied by 5 dpi. Macroscopic lesions were recorded, and swabs and fragments of thoracic and abdominal organs were analyzed by bacteriological and pathological assays. The PmA strains were analyzed for four virulence genes (toxA: toxin; pfhA: adhesion; tbpA and hgbB: iron acquisition) by PCR and sequencing and submitted to multilocus sequence typing (MLST).
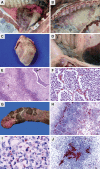
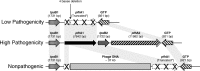
Results: The eight PmA strains were classified as follows: five as highly pathogenic (HP) for causing necrotic bronchopneumonia and diffuse fibrinous pleuritis and pericarditis; one as low pathogenic for causing only focal bronchopneumonia; and two as nonpathogenic because they did not cause injury to any pig. PCR for the gene pfhA was positive for all five HP isolates. Sequencing demonstrated that the pfhA region of the HP strains comprised four genes: tpsB1, pfhA1, tpsB2 and pfhA2. The low and nonpathogenic strains did not contain the genes tpsB2 and pfhA2. A deletion of four bases was observed in the pfhA gene in the low pathogenic strain, and an insertion of 37 kb of phage DNA was observed in the nonpathogenic strains. MLST clustered the HP isolates in one group and the low and nonpathogenic isolates in another. Only the nonpathogenic isolates matched sequence type 10; the other isolates did not match any type available in the MLST database.
Conclusions: The hypothesis that some PmA strains are primary pathogens and cause disease in pigs without any co-factor was confirmed. The pfhA region, comprising the genes tpsB1, tpsB2, pfhA1 and pfhA2, is related to the pathogenicity of PmA. The HP strains can cause necrotic bronchopneumonia, fibrinous pleuritis and pericarditis in pigs and can be identified by PCR amplification of the gene pfhA2.
Keywords: Bronchopneumonia; MLST; Pigs; Polyserositis; Respiratory diseases; pfhA.
Conflict of interest statement
Ethics approval
The experiment was conducted at the Embrapa Swine and Poultry Research Center, Concordia-SC, Brazil, in compliance with the Ethical Principles in Animal Experimentation adopted by the National Council for Control of Animal Experimentation (CONCEA) and approved by the Ethics Committee on Animal Experimentation (CEUA/CNPSA) (Protocol #005/2010). All the experimental procedures were performed strictly in accordance with the approved guidelines and regulations of Institutional Animal Ethics Committee.
Consent for publication
Not applicable in these sections.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Characterization of Pasteurella multocida associated with ovine pneumonia using multi-locus sequence typing (MLST) and virulence-associated gene profile analysis and comparison with porcine isolates.Vet Microbiol. 2017 May;204:180-187. doi: 10.1016/j.vetmic.2017.04.015. Epub 2017 Apr 22. Vet Microbiol. 2017. PMID: 28532799
-
A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China.Arch Microbiol. 2018 Jan;200(1):107-118. doi: 10.1007/s00203-017-1421-y. Epub 2017 Aug 20. Arch Microbiol. 2018. PMID: 28825122
-
Pasteurella multocida polyserositis in growing-finishing pigs.J Comp Pathol. 2023 Apr;202:16-22. doi: 10.1016/j.jcpa.2023.03.003. Epub 2023 Apr 4. J Comp Pathol. 2023. PMID: 37023584
-
[Pasteurella: insights into the virulence determinants of a heterogenous bacterial type].Berl Munch Tierarztl Wochenschr. 2004 Sep-Oct;117(9-10):367-86. Berl Munch Tierarztl Wochenschr. 2004. PMID: 15495927 Review. German.
-
[Molecular typing methods for Pasteurella multocida-A review].Wei Sheng Wu Xue Bao. 2016 Oct 4;56(10):1521-9. Wei Sheng Wu Xue Bao. 2016. PMID: 29741340 Review. Chinese.
Cited by
-
Comparative analysis reveals the Genomic Islands in Pasteurella multocida population genetics: on Symbiosis and adaptability.BMC Genomics. 2019 Jan 18;20(1):63. doi: 10.1186/s12864-018-5366-6. BMC Genomics. 2019. PMID: 30658579 Free PMC article.
-
Effect of Vaccination against Glässer's Disease in a Farm Suffering from Polyserositis in Weaned Pigs.Vet Sci. 2022 Dec 12;9(12):691. doi: 10.3390/vetsci9120691. Vet Sci. 2022. PMID: 36548852 Free PMC article.
-
18β-Glycyrrhetinic Acid Alleviates P. multocida-Induced Vascular Endothelial Inflammation by PARP1-Mediated NF-κB and HMGB1 Signalling Suppression in PIEC Cells.Infect Drug Resist. 2023 Jun 29;16:4201-4212. doi: 10.2147/IDR.S413242. eCollection 2023. Infect Drug Resist. 2023. PMID: 37404255 Free PMC article.
-
In vitro and in vivo Synergistic Effects of Florfenicol and Thiamphenicol in Combination Against Swine Actinobacillus pleuropneumoniae and Pasteurella multocida.Front Microbiol. 2019 Oct 30;10:2430. doi: 10.3389/fmicb.2019.02430. eCollection 2019. Front Microbiol. 2019. PMID: 31749775 Free PMC article.
-
Development of an Optimal Model of Combined Radiation and Biological Lesions.Vet Med Int. 2022 Sep 8;2022:9433032. doi: 10.1155/2022/9433032. eCollection 2022. Vet Med Int. 2022. PMID: 36118594 Free PMC article.
References
-
- Pijoan C, Fuentes M. Severe pleuritis associated with certain strains of Pasteurella multocida in swine. J Am Vet Med Assoc. 1987;191:823–826. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

